# A tibble: 32,735 × 16
year month day dep_time dep_delay arr_time arr_delay carrier tailnum
<int> <int> <int> <int> <dbl> <int> <dbl> <chr> <chr>
1 2013 6 30 940 15 1216 -4 VX N626VA
2 2013 5 7 1657 -3 2104 10 DL N3760C
3 2013 12 8 859 -1 1238 11 DL N712TW
4 2013 5 14 1841 -4 2122 -34 DL N914DL
5 2013 7 21 1102 -3 1230 -8 9E N823AY
6 2013 1 1 1817 -3 2008 3 AA N3AXAA
7 2013 12 9 1259 14 1617 22 WN N218WN
8 2013 8 13 1920 85 2032 71 B6 N284JB
9 2013 9 26 725 -10 1027 -8 AA N3FSAA
10 2013 4 30 1323 62 1549 60 EV N12163
# ℹ 32,725 more rows
# ℹ 7 more variables: flight <int>, origin <chr>, dest <chr>, air_time <dbl>,
# distance <dbl>, hour <dbl>, minute <dbl>Environmental Data Analysis and Visualization
A brief reminder
This course deals with the representation of data as a means to communicating about important issues. Our ability to consider a wide and topical range of data representations depends on respect for each other and a commitment to our shared motivation to learn.
Visualization Critique
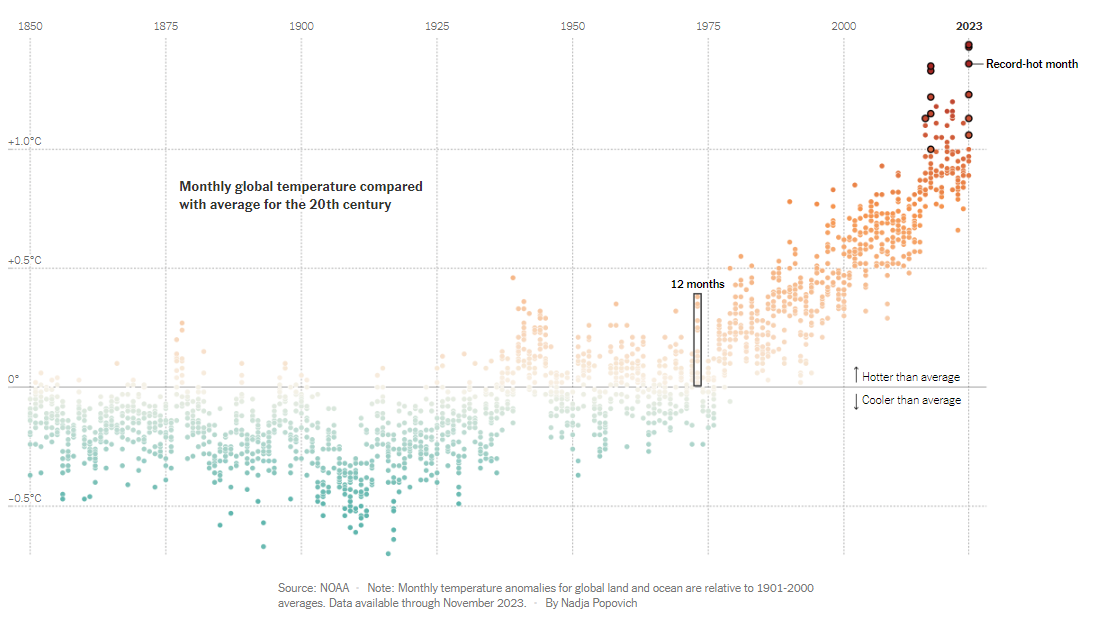
Visualization Critique
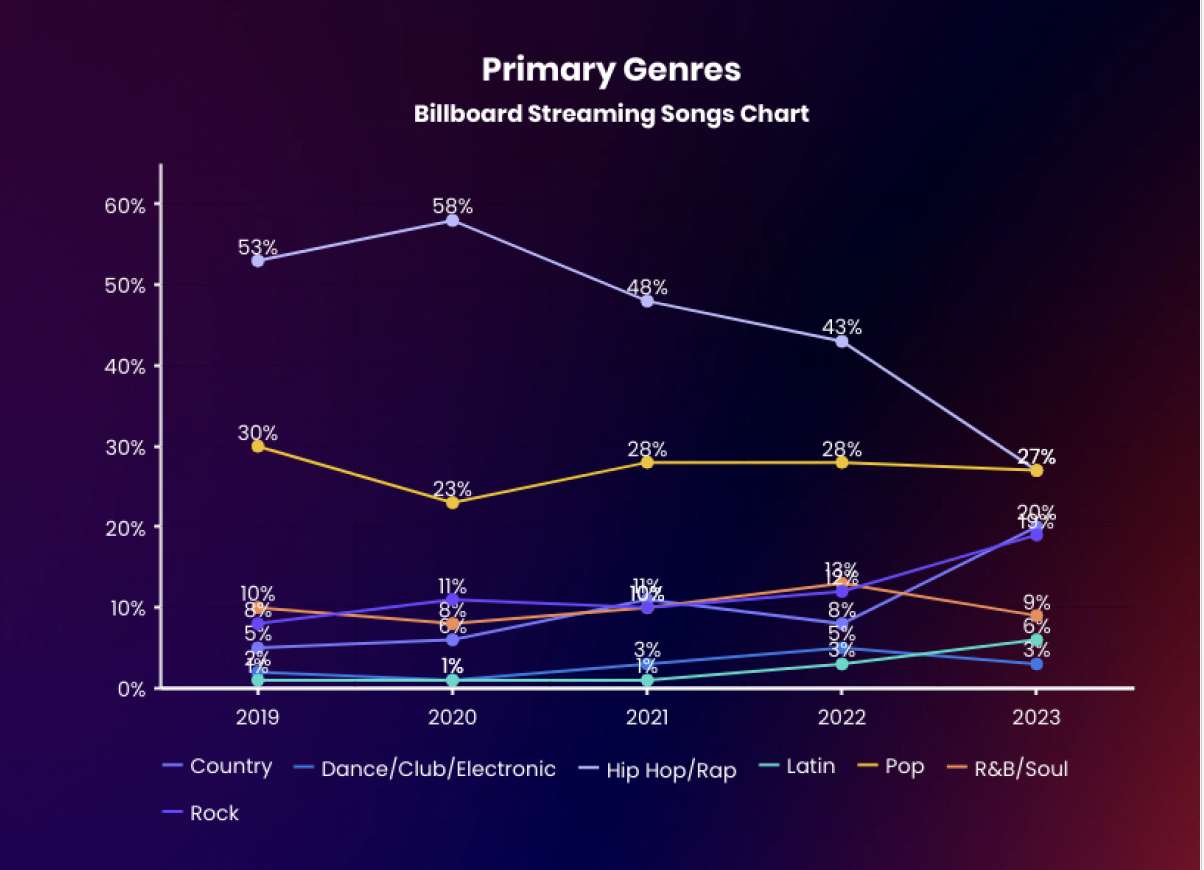
Visualization Critique
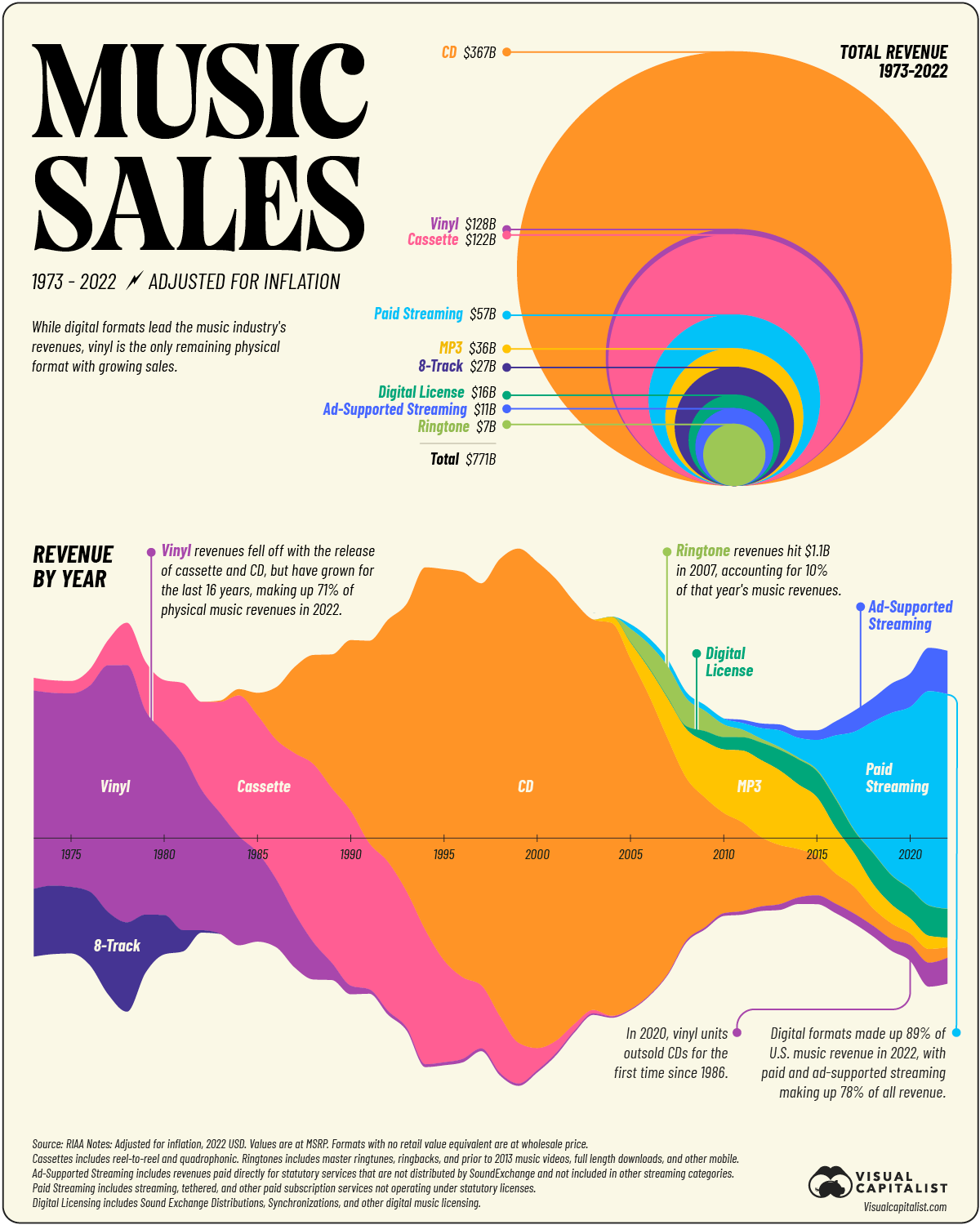
Visualization Critique
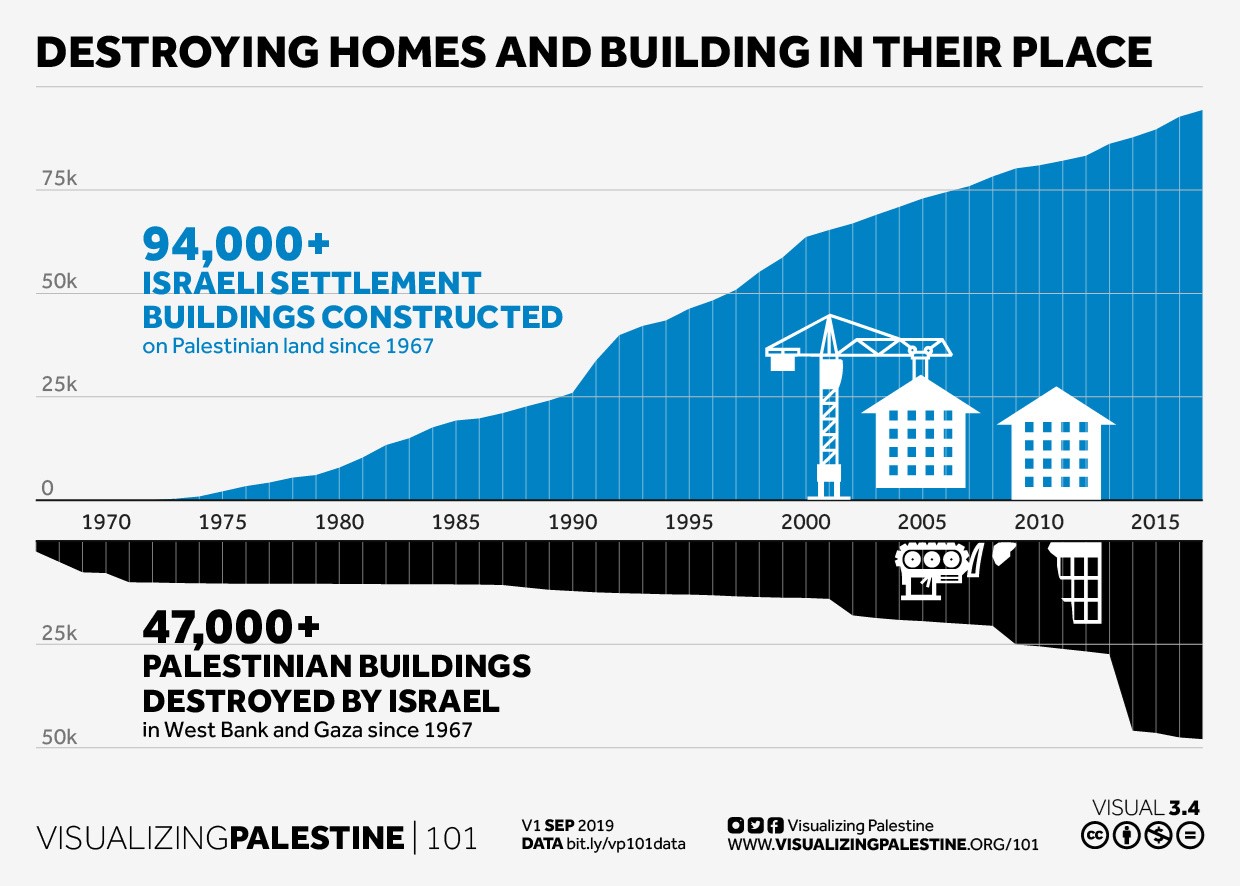
Next week’s critiques
Mark
Jenny
Mia
Data
Data are individual pieces of information.

Datasets
Data in the field

Data in the field
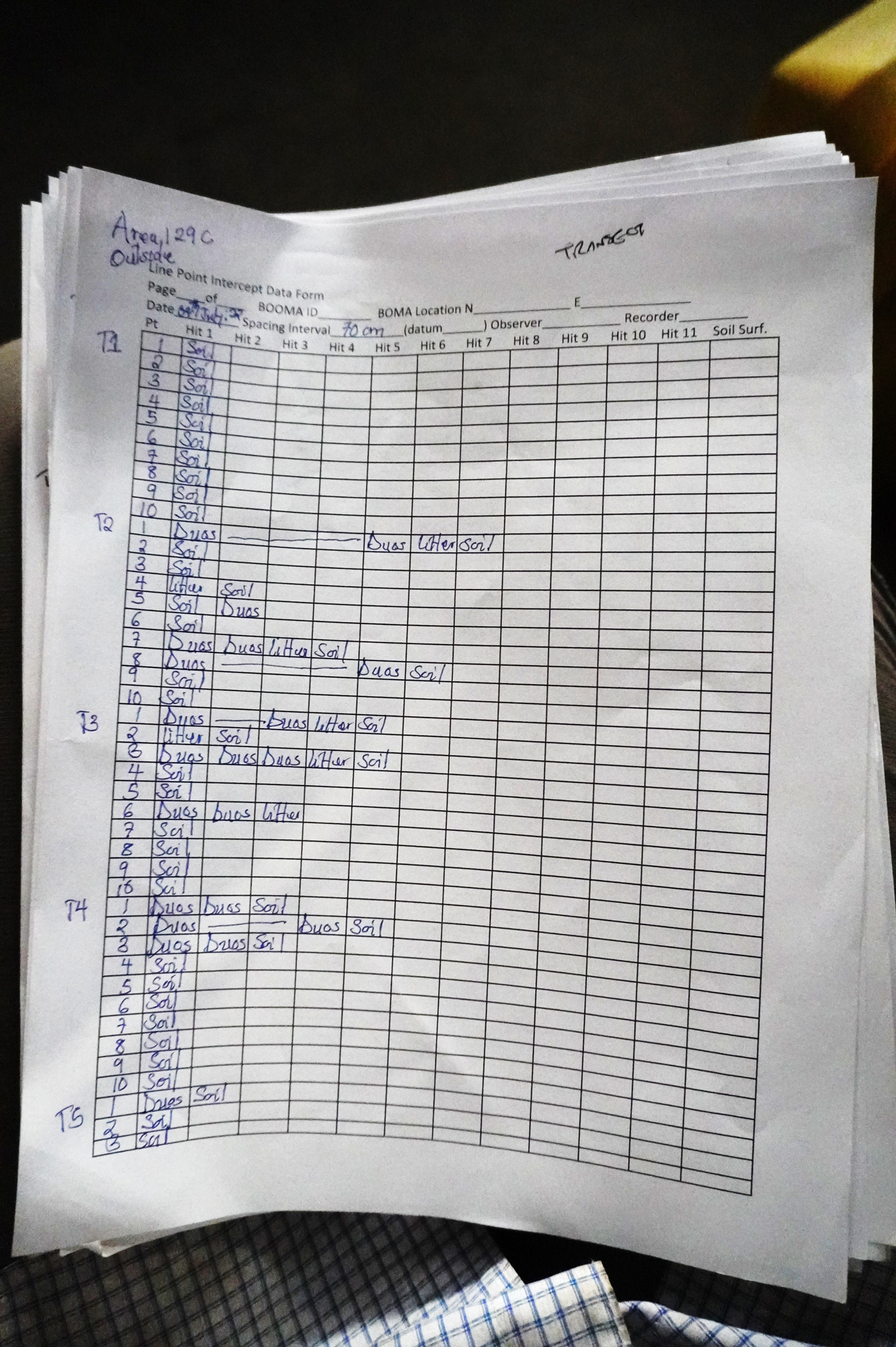
Messy data
The process of data entry is often designed and conducted with limited consideration given to the process of data analysis.

https://openscapes.org/blog/2020-10-12-tidy-data/
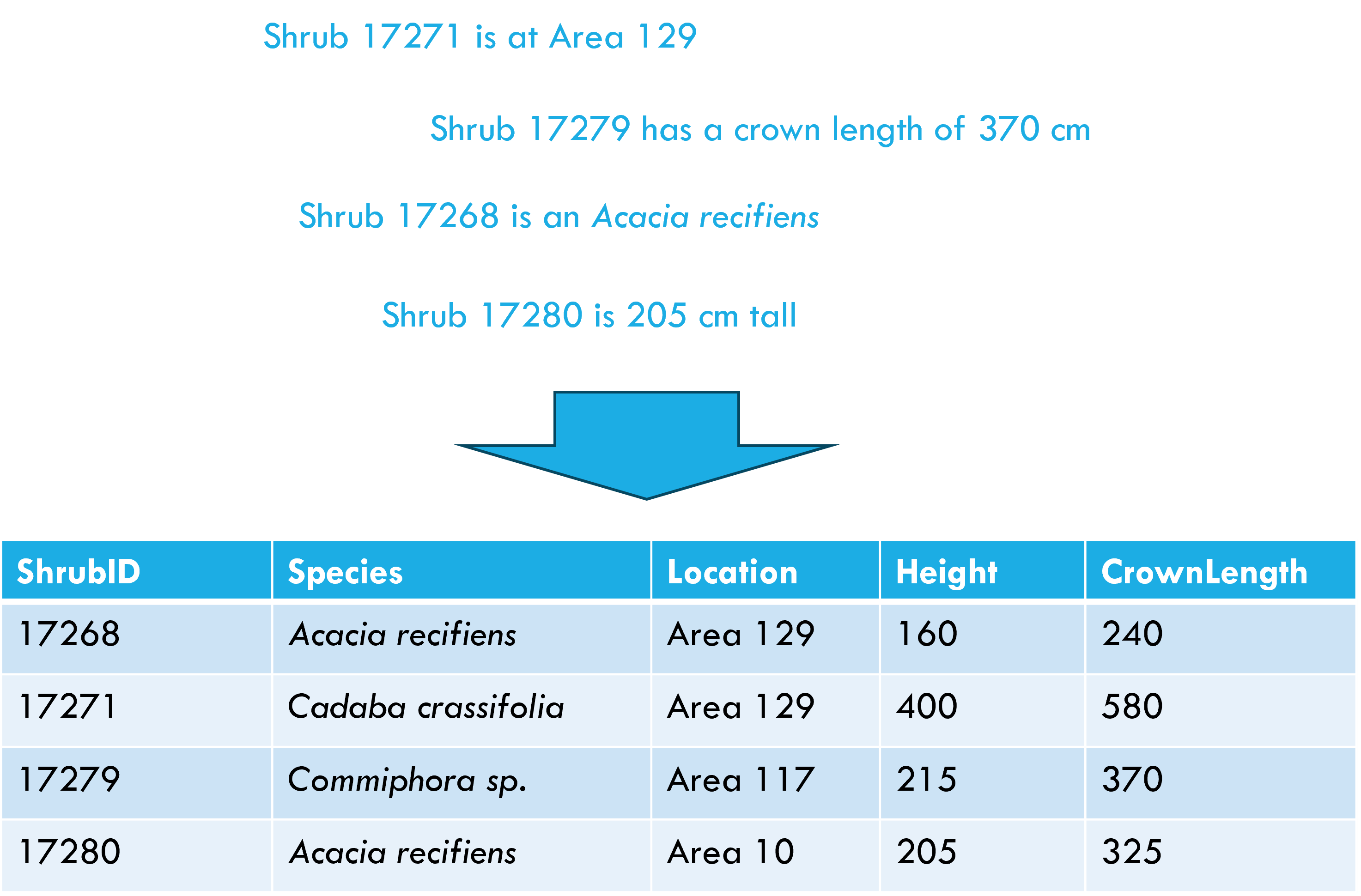
Tidy data principles
Each variable forms a column
Each observation forms a row
Each cell contains a single value
After Wickham, H. 2014. Tidy data. The Journal of Statistical Software 59.
Vectorization

Querying


Consistency

Ease of identification and re-use

Data wrangling: the big three
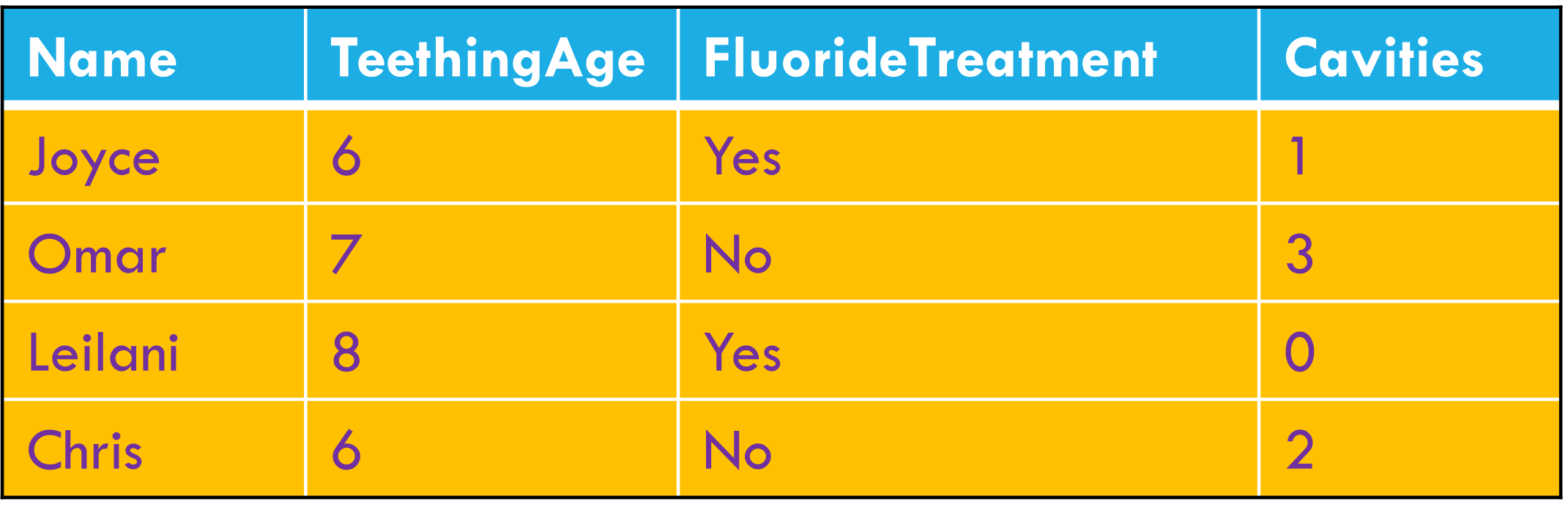
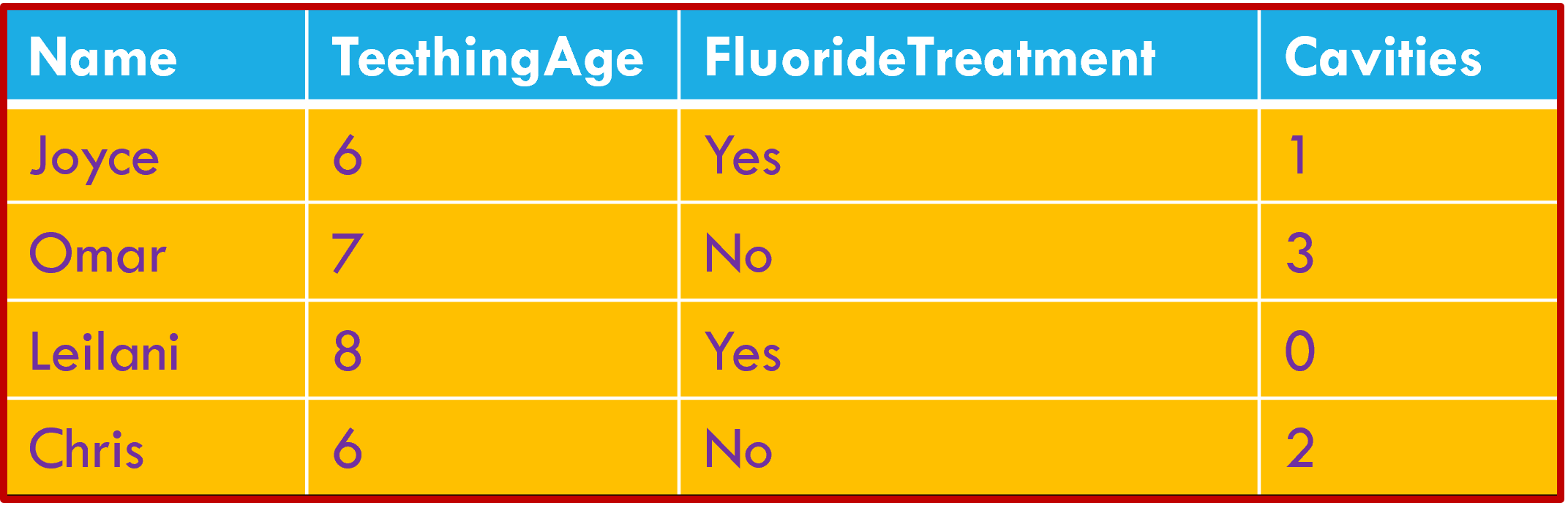
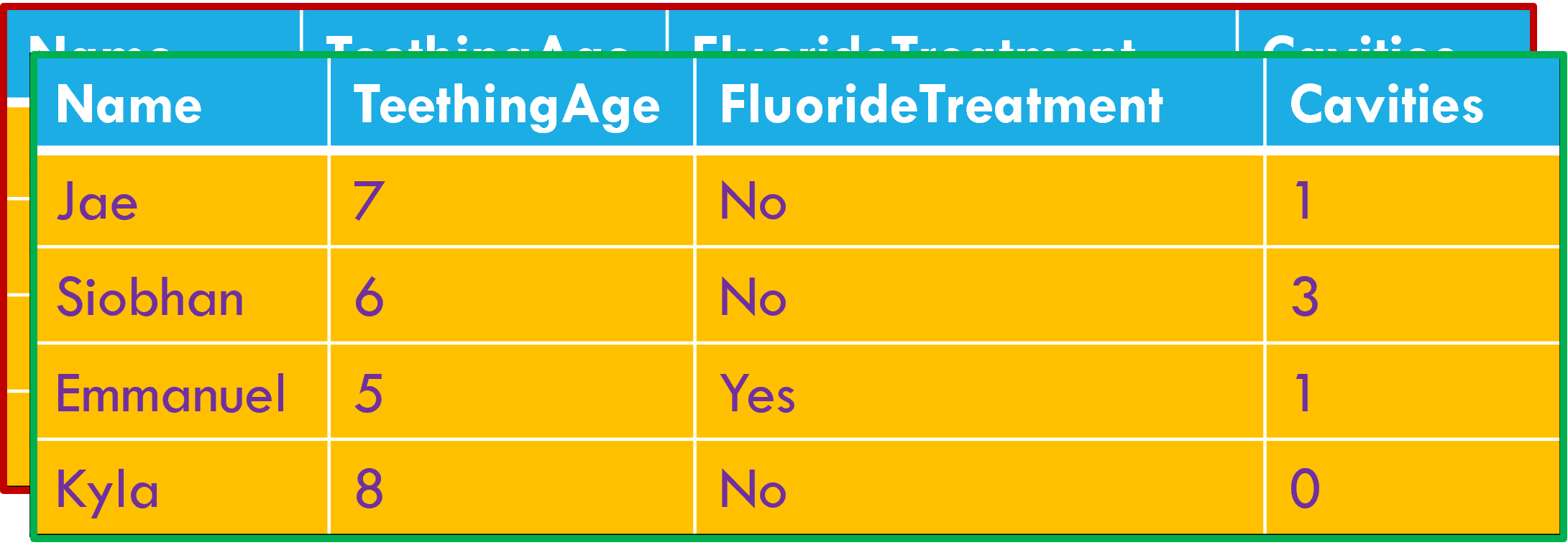
Column headers are values, not variable names.
Data wrangling: the big three
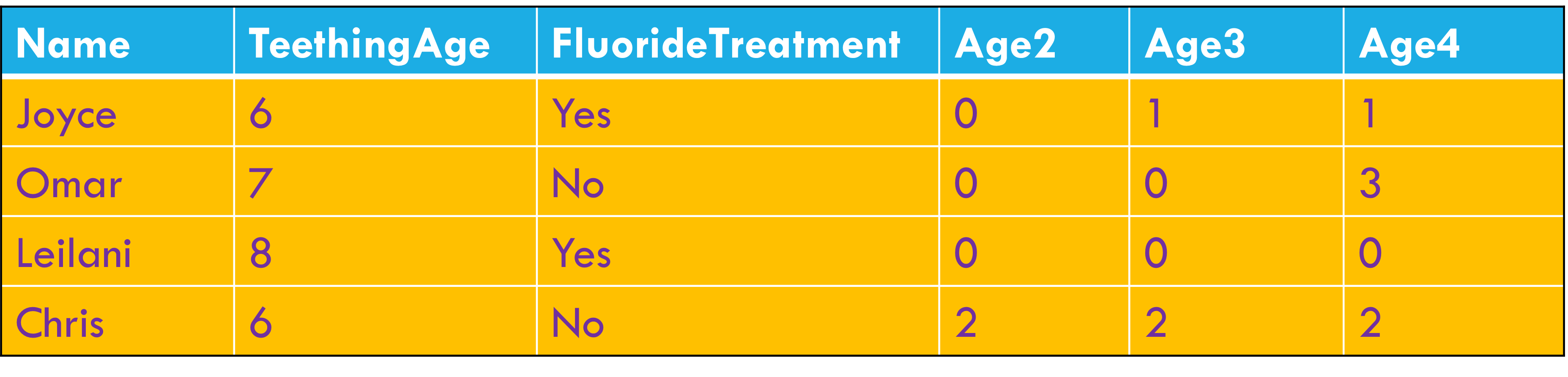
Multiple variables are stored in a single column.
Data wrangling: the big three
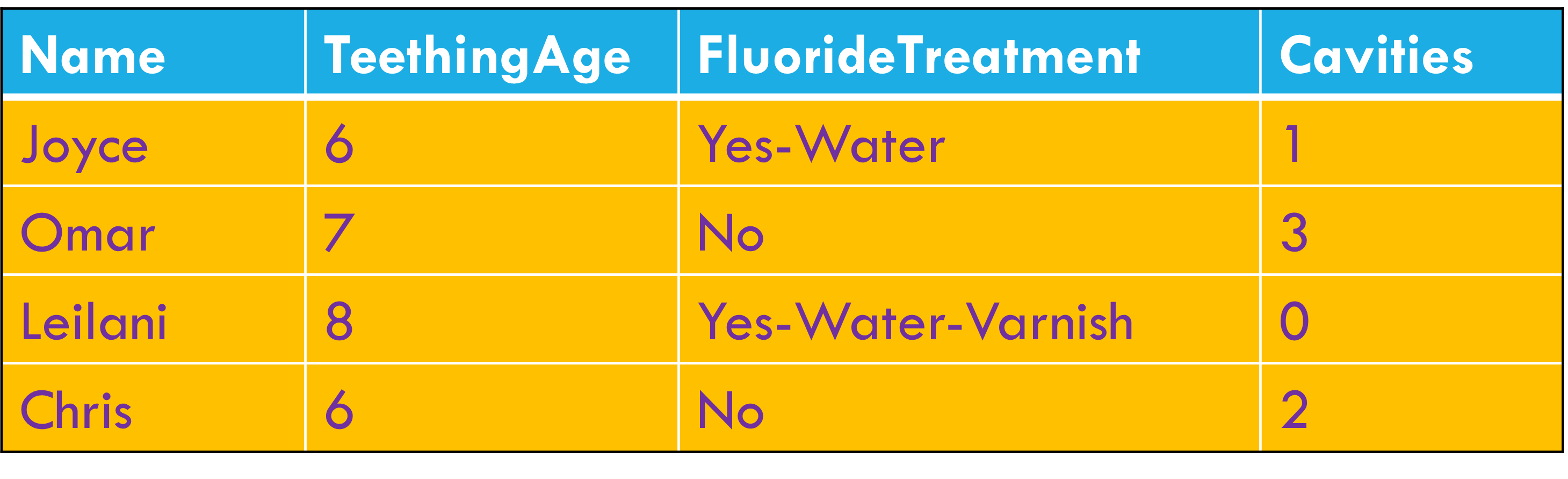
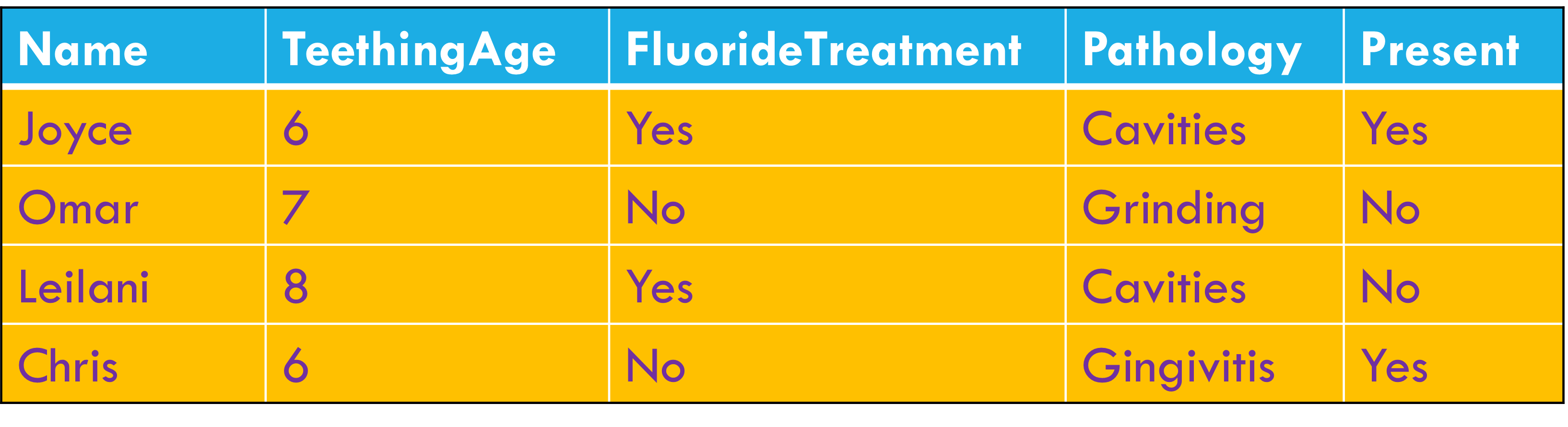
Variables stored in both rows and columns.
Pipes in R
The pipe operator %>% allows us to combine multiple functions into an ordered set of transformations
Pipes in R
Pipes in R
# A tibble: 2,610 × 16
year month day dep_time dep_delay arr_time arr_delay carrier tailnum
<int> <int> <int> <int> <dbl> <int> <dbl> <chr> <chr>
1 2013 1 1 1817 -3 2008 3 AA N3AXAA
2 2013 1 23 2024 37 2141 29 EV N17115
3 2013 1 15 1626 -3 1941 10 B6 N594JB
4 2013 1 17 626 -4 846 3 US N554UW
5 2013 1 8 902 -3 1006 -17 B6 N281JB
6 2013 1 15 1947 167 2241 171 AA N5EGAA
7 2013 1 1 1454 -4 1554 -21 EV N11544
8 2013 1 30 1306 -9 1430 -1 EV N13969
9 2013 1 4 1942 -3 2249 -40 B6 N637JB
10 2013 1 8 1859 -6 2158 -27 AA N322AA
# ℹ 2,600 more rows
# ℹ 7 more variables: flight <int>, origin <chr>, dest <chr>, air_time <dbl>,
# distance <dbl>, hour <dbl>, minute <dbl>Data pipelines
Pipelines start with raw data (e.g., tibble) as the input
This table is passed as the first argument to the next function (so they don’t need the data argument)
The result of a pipeline is a dataset that can be fed into visualizations and analysis.
Data pipelines
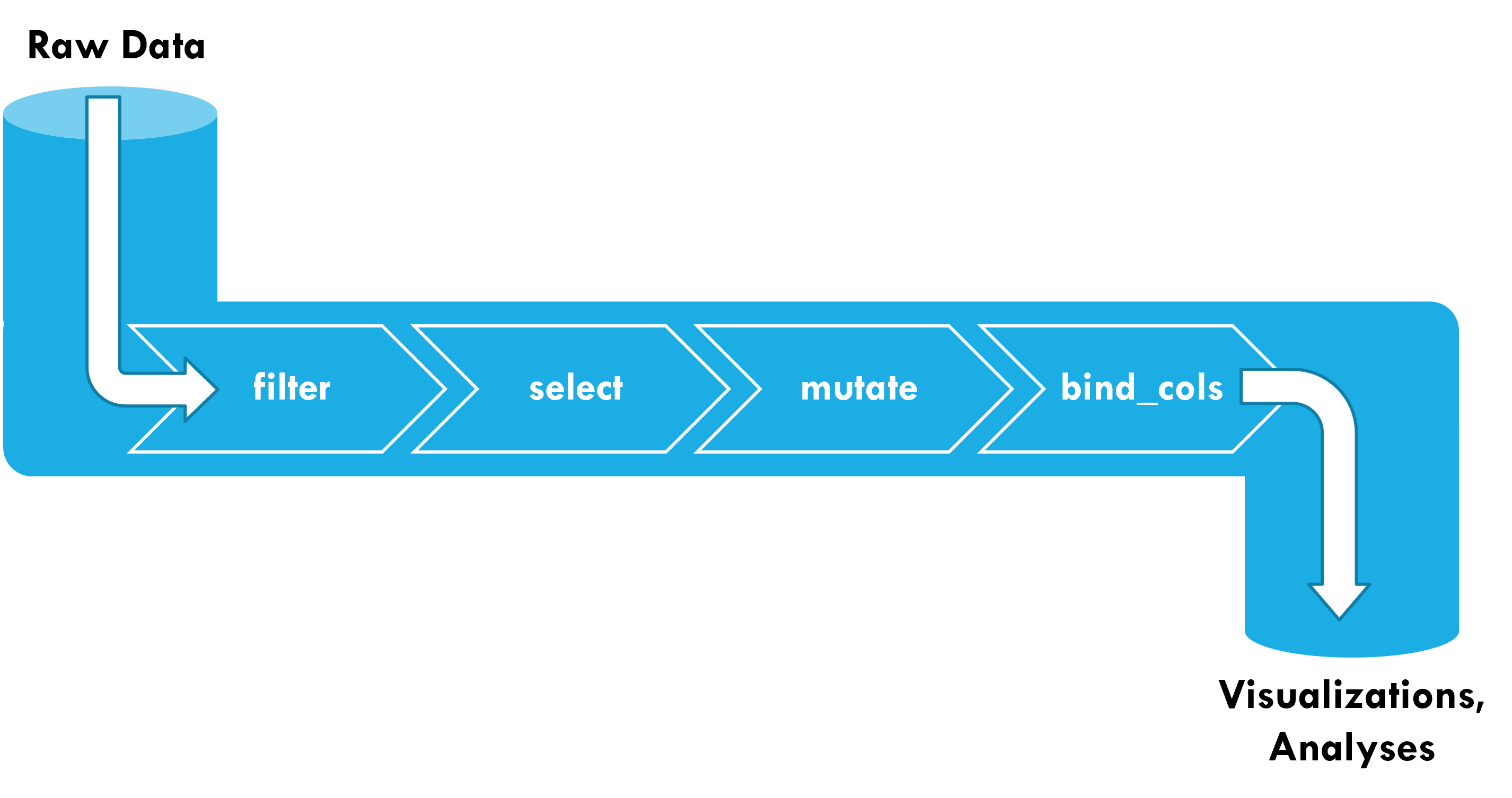
Data pipelines
nycflights %>%
mutate(speed = distance / air_time * 60) %>%
select(year:day,dep_time,carrier,flight,speed) %>%
filter(month==1) %>%
unite(col="Date",c(month,day,year),sep="/")# A tibble: 2,610 × 5
Date dep_time carrier flight speed
<chr> <int> <chr> <int> <dbl>
1 1/1/2013 1817 AA 353 319.
2 1/23/2013 2024 EV 4412 319.
3 1/15/2013 1626 B6 369 414
4 1/17/2013 626 US 1433 311.
5 1/8/2013 902 B6 56 340.
6 1/15/2013 1947 AA 575 396.
7 1/1/2013 1454 EV 4390 363.
8 1/30/2013 1306 EV 4120 277.
9 1/4/2013 1942 B6 645 466.
10 1/8/2013 1859 AA 21 441.
# ℹ 2,600 more rowsActivity: Going with the directional movement of a fluid
Working in pairs or small groups, use the different tidyverse functions to create a pipeline to wrangle raw data into the desired form
Datasets will come from Canvas and the modeldata package
Use cheatsheets on Canvas for function descriptions
Example 1
Body mass and flipper length for penguins from Biscoe and Dream islands with a body mass over 400 grams.
Example 2
Price, number of baths, and square meters for three-bedroom homes in the Roseville, Orangevale, and Citrus Heights neighborhoods of Sacramento.
Example 3
ID Number, Diameter in mm, and number of rings on top 100 abalone by ring count.
Example 3
rawData<-read_csv("data/abalone.csv")
rawData %>%
mutate(diameter_mm=diameter * 200) %>%
select(...1,diameter_mm,rings) %>%
slice_max(order_by=rings,n=100)# A tibble: 136 × 3
...1 diameter_mm rings
<dbl> <dbl> <dbl>
1 481 117 29
2 2109 107 27
3 2210 93 27
4 295 99 26
5 2202 98 25
6 3150 108 24
7 3281 108 24
8 314 94 23
9 315 97 23
10 502 104 23
# ℹ 126 more rowsCan I use pipes with ggplot2?
Can I use pipes with ggplot2?
YES!
Can I use pipes with Base R?
Can I use pipes with Base R?
YES!
Can I use pipes with Base R?
YES!*
Can I use pipes with Base R?
YES!*
rawData %>%
mutate(wholeWeight=weight.whole * 200) %>%
mutate(shuckedWeight=weight.shucked * 200) %>%
select(wholeWeight,shuckedWeight) %>%
{sum(.$wholeWeight)}[1] 692331.2*certain rules apply when first argument is not the first argument, or when referencing column names
Can I use pipes for modeling data?
YES!
rawData %>%
mutate(diameter_mm=diameter * 200) %>%
select(...1,diameter_mm,rings) %>%
lm(rings ~ diameter_mm, data=.) %>%
summary()
Call:
lm(formula = rings ~ diameter_mm, data = .)
Residuals:
Min 1Q Median 3Q Max
-5.1868 -1.6932 -0.7200 0.9066 15.9999
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 2.318574 0.172737 13.42 <2e-16 ***
diameter_mm 0.093350 0.002057 45.37 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 2.639 on 4175 degrees of freedom
Multiple R-squared: 0.3302, Adjusted R-squared: 0.3301
F-statistic: 2059 on 1 and 4175 DF, p-value: < 2.2e-16Coursekeeping
Coding exercise 2 is now available on Canvas, due November 6!

the-decoder.com
Next week
Visualizing more relationships
Fine controls with
ggplot2Time is weird