library(tidyverse)Zebra Teeth Simulation Outcomes
Load packages
Birth and mortality profiles
Birth
birthDeath<-read_csv("birthDeath.csv")
birthDeath$born<-month.abb[birthDeath$born]
ggplot(birthDeath,aes(x=fct_relevel(as_factor(born),"Jan","Feb","Mar","Apr","May","Jun","Jul","Aug","Sep","Oct","Nov","Dec"))) +
geom_bar(fill="darkblue") +
theme_classic() +
labs(x="Birth month",y="n") +
theme(
axis.text=element_text(size=12),
axis.title=element_text(size=14),
axis.text.x = element_text(angle = 45,hjust=1))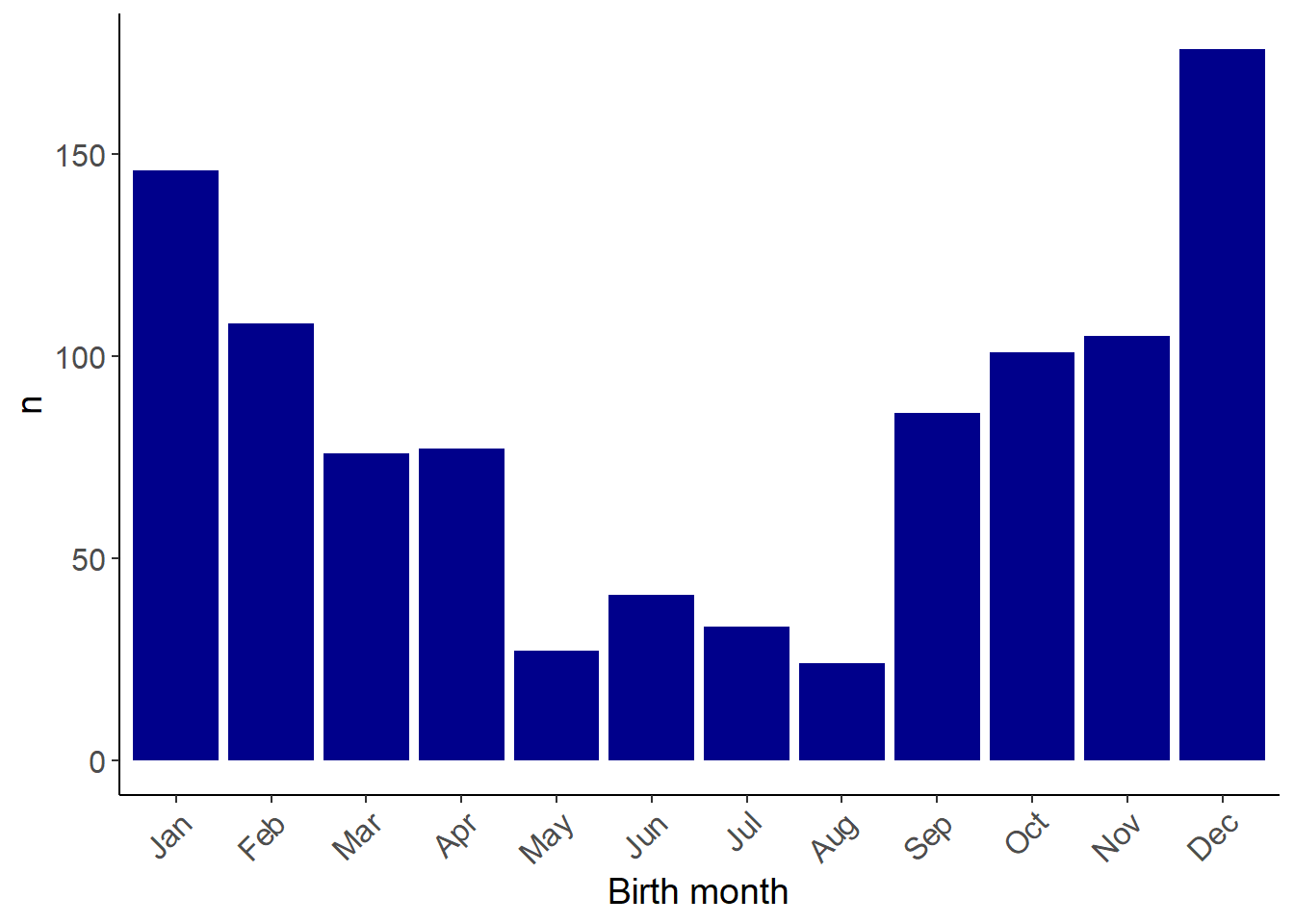
Death
ggplot(birthDeath,aes(x=died)) +
geom_histogram(fill="darkred") +
theme_classic() +
labs(x="Age at death (years)",y="n") +
theme(
axis.text=element_text(size=12),
axis.title=element_text(size=14))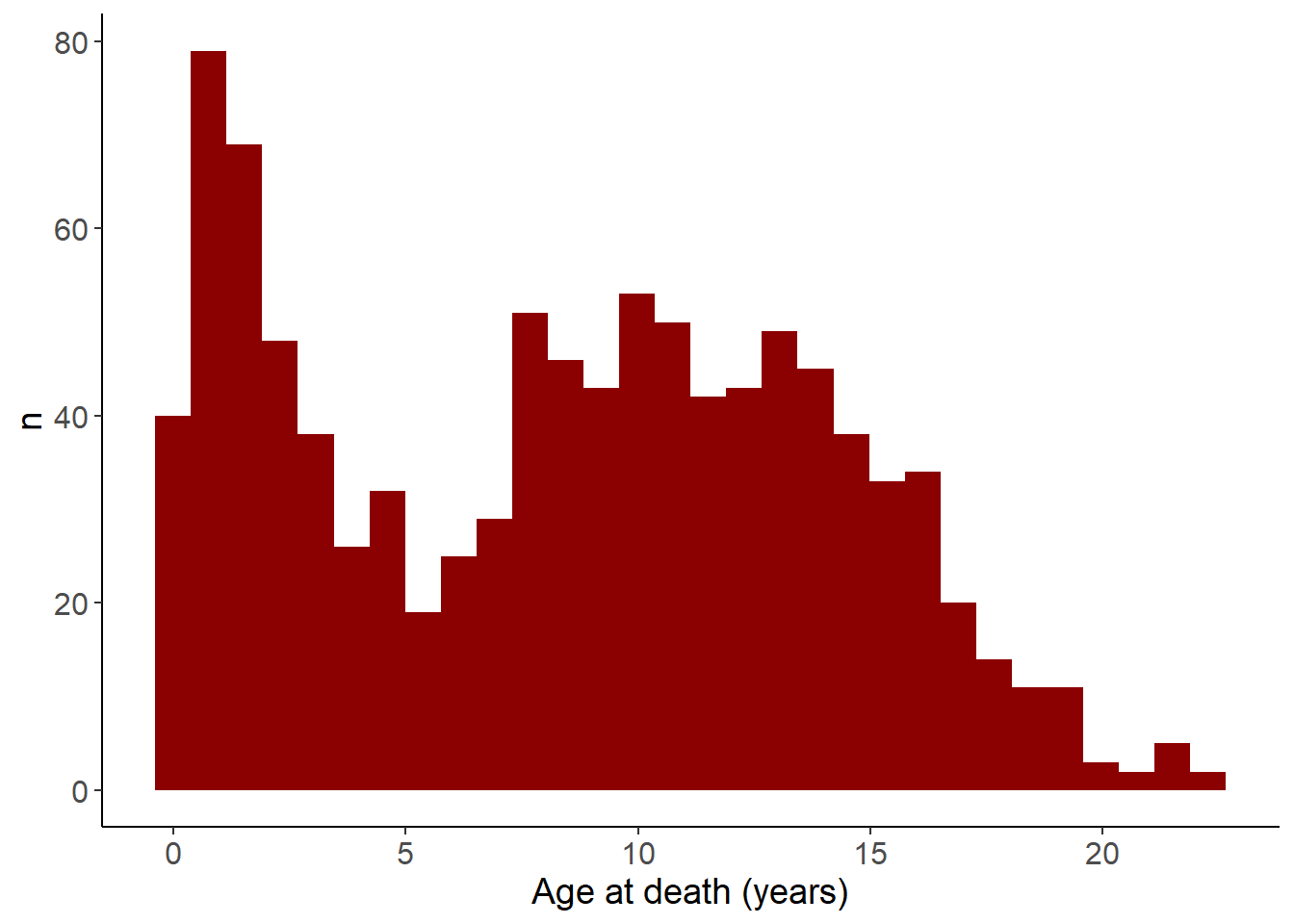
Tooth growth models
toothGrowth<-read_csv("growthRates.csv") %>%
pivot_longer(cols=constant:nonLinear,names_to="Model",values_to="crownHeight")
ggplot(toothGrowth,aes(x=day,y=crownHeight,color=fct_recode(Model,Constant="constant","Non-linear"="nonLinear"))) +
geom_line(lwd=1.1) +
scale_color_manual(values=c("black","darkorange")) +
theme_classic() +
labs(x="Days since start of mineralization",y="Crown Height (\u{03BC}m)",color="Model") +
theme(axis.text=element_text(size=12),axis.title=element_text(size=14))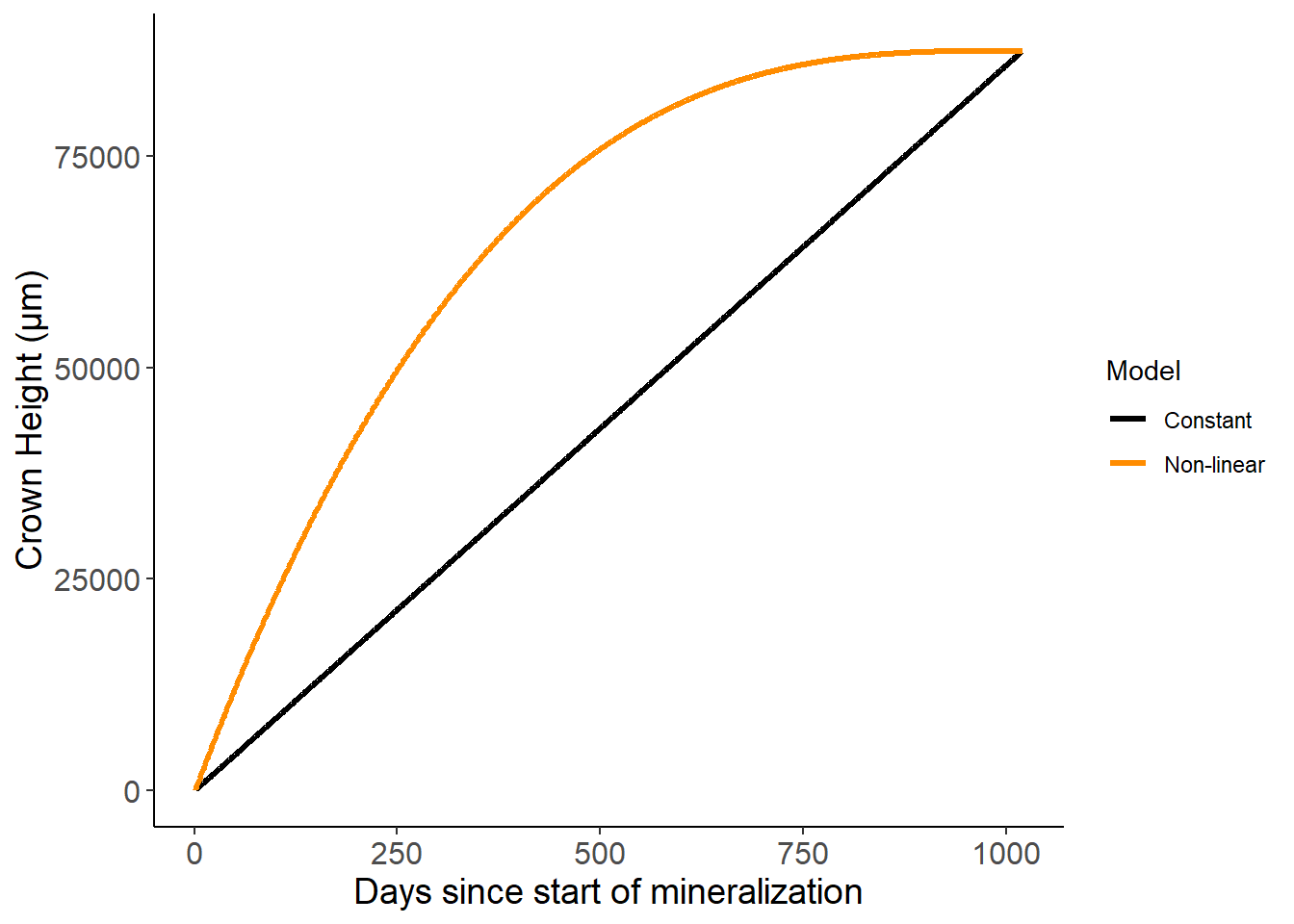
Model 𝝳18O signals
Amplitudes of 1, 2, 3, and 4‰ were simulated; most results below are based on the 3‰ signal.
d18O<-read_csv("sim_d18O.csv") %>%
pivot_longer(cols=AMP1:AMP4,names_to="Amplitude",values_to="d18O")
ggplot(d18O,aes(x=as_date(DAY,origin="2000-12-21"),y=d18O,color=case_match(Amplitude,"AMP1"~"1","AMP2"~"2","AMP3"~"3","AMP4"~"4"))) +
geom_hline(yintercept=-4,linetype=2)+
geom_line(lwd=1.1) +
scale_color_manual(values=palette.colors(4,palette="R4")) +
scale_x_date(date_labels = "%b", date_breaks = '3 months') +
theme_classic() +
labs(x="",y=expression(paste(delta^"18","O")),color="Amplitude (\u{2030})") +
theme(axis.text=element_text(size=12),axis.title=element_text(size=14))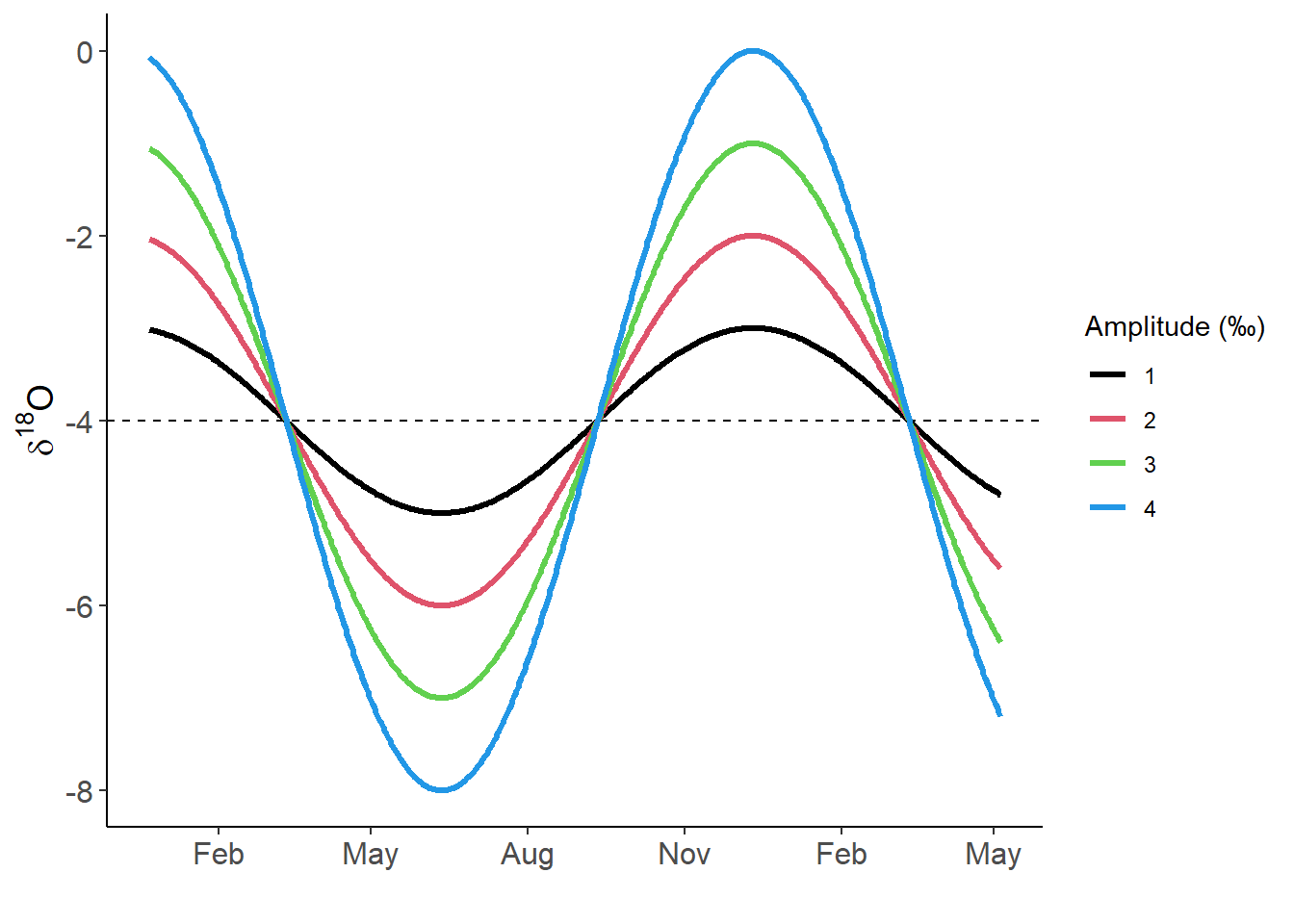
Constant vs non-linear growth rate (no wear; Amplitude = 3)
These teeth come from animals that are all born on the same day (Jan 1) and die after 55 months.
Just the constant model:
Ex1<-read_csv("simZebraDataEX12.csv") %>% pivot_longer(cols=d18O_1:d18O_4,names_to="Amplitude",values_to="d18O",names_prefix="d18O_")
Ex1_Amp3<-filter(Ex1,Amplitude==3,specimen==1)
Ex1_Amp3_con<-filter(Ex1_Amp3,model=="Constant")
ggplot(Ex1_Amp3_con,aes(x=mm_band,y=d18O)) +
geom_line() +
geom_hline(yintercept=-4,linetype=2)+
geom_line(lwd=1.1) +
theme_classic() +
labs(x="Distance from Occlusal Surface (mm)",y=expression(paste(delta^"18","O")))+
theme(axis.text=element_text(size=12),axis.title=element_text(size=14))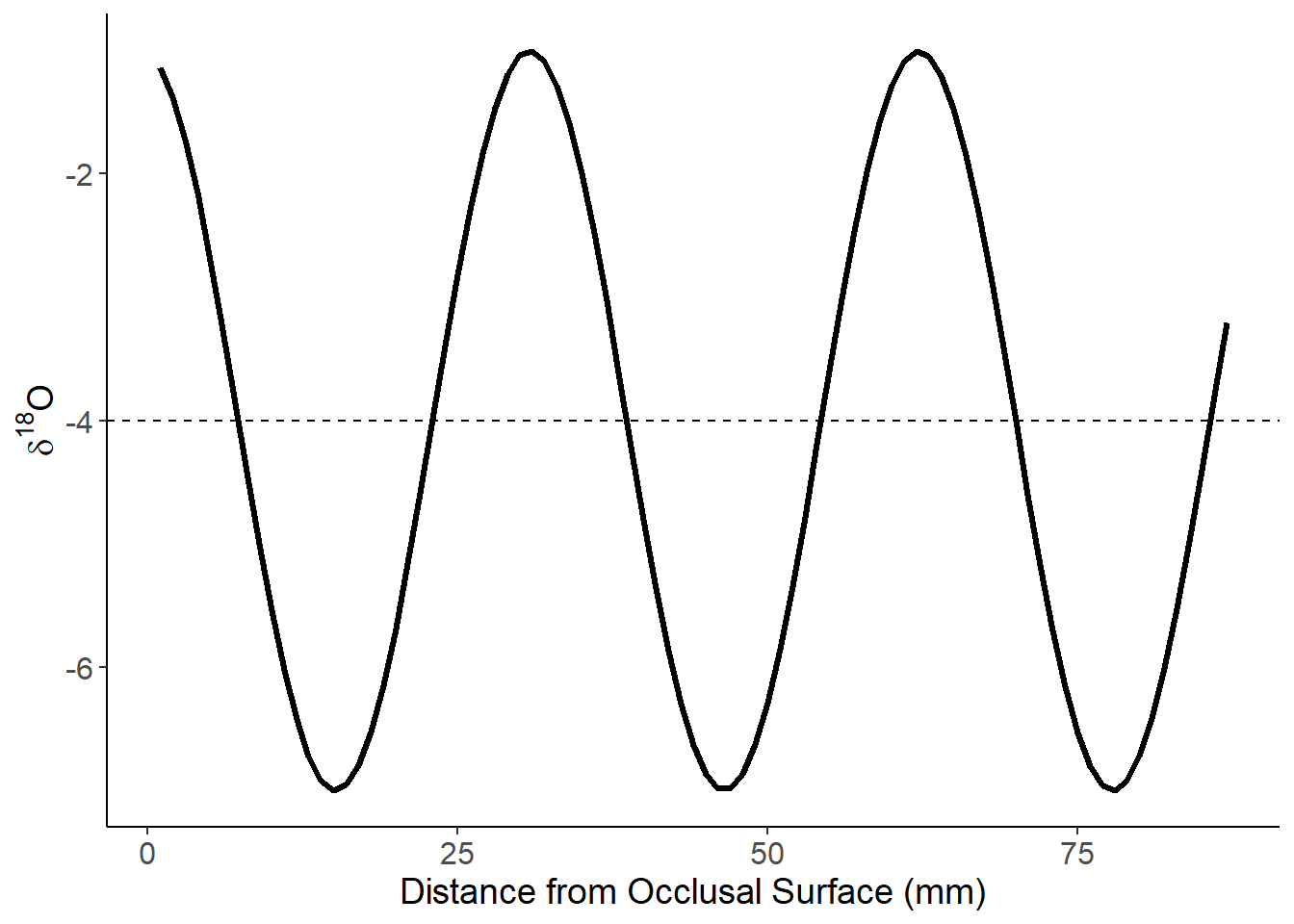
Constant and non-linear
ggplot(Ex1_Amp3,aes(x=mm_band,y=d18O,color=fct_recode(model,"Non-Linear" = "High"))) + geom_line() +
geom_hline(yintercept=-4,linetype=2)+
geom_line(lwd=1.1) +
scale_color_manual(values=c("black","darkorange")) +
theme_classic() +
labs(x="Distance from Occlusal Surface (mm)",y=expression(paste(delta^"18","O")),color="Model")+
theme(axis.text=element_text(size=12),axis.title=element_text(size=14))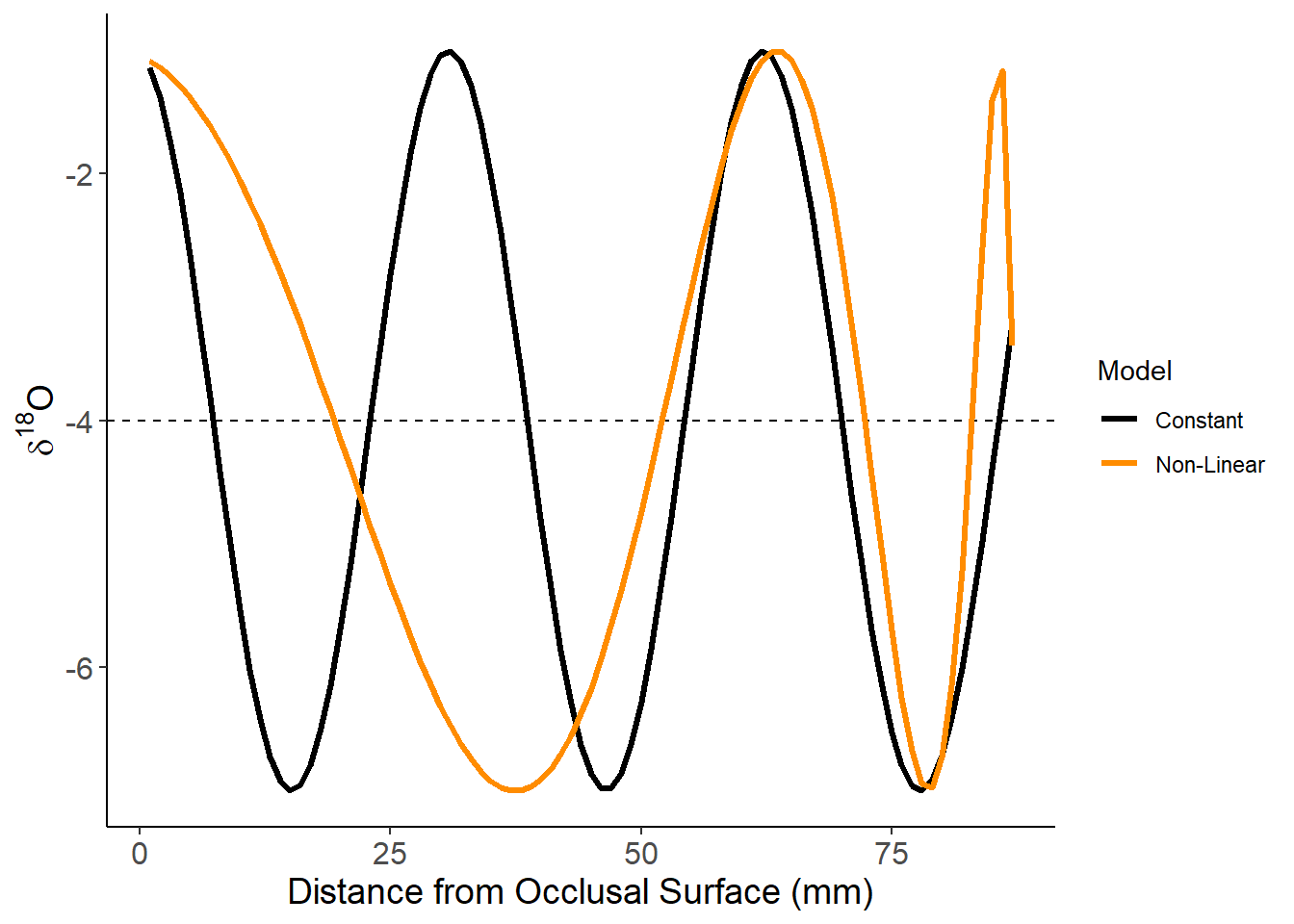
Non-linear growth with variable birth season ( Amplitude = 3)
Ex3<-read_csv("simZebraDataEX3.csv") %>% pivot_longer(cols=d18O_1:d18O_4,names_to="Amplitude",values_to="d18O",names_prefix="d18O_")Rows: 55000 Columns: 17
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (1): model
dbl (16): specimen, age_at_death, birth_year, birth_month, birth_day, crown_...
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.uv<-unique(Ex3$specimen)
samp<-sample(uv,50,replace=FALSE)
Ex3_Amp3<-filter(Ex3,Amplitude==3,model=="High", specimen %in% samp)
ggplot(Ex3_Amp3,aes(x=mm_band,y=d18O,group=specimen,color=specimen)) +
geom_hline(yintercept=-4,linetype=2)+
geom_line(alpha=0.8,color="grey") +
theme_classic() +
labs(x="Distance from Occlusal Surface (mm)",y=expression(paste(delta^"18","O"))) +
theme(axis.text=element_text(size=12),axis.title=element_text(size=14))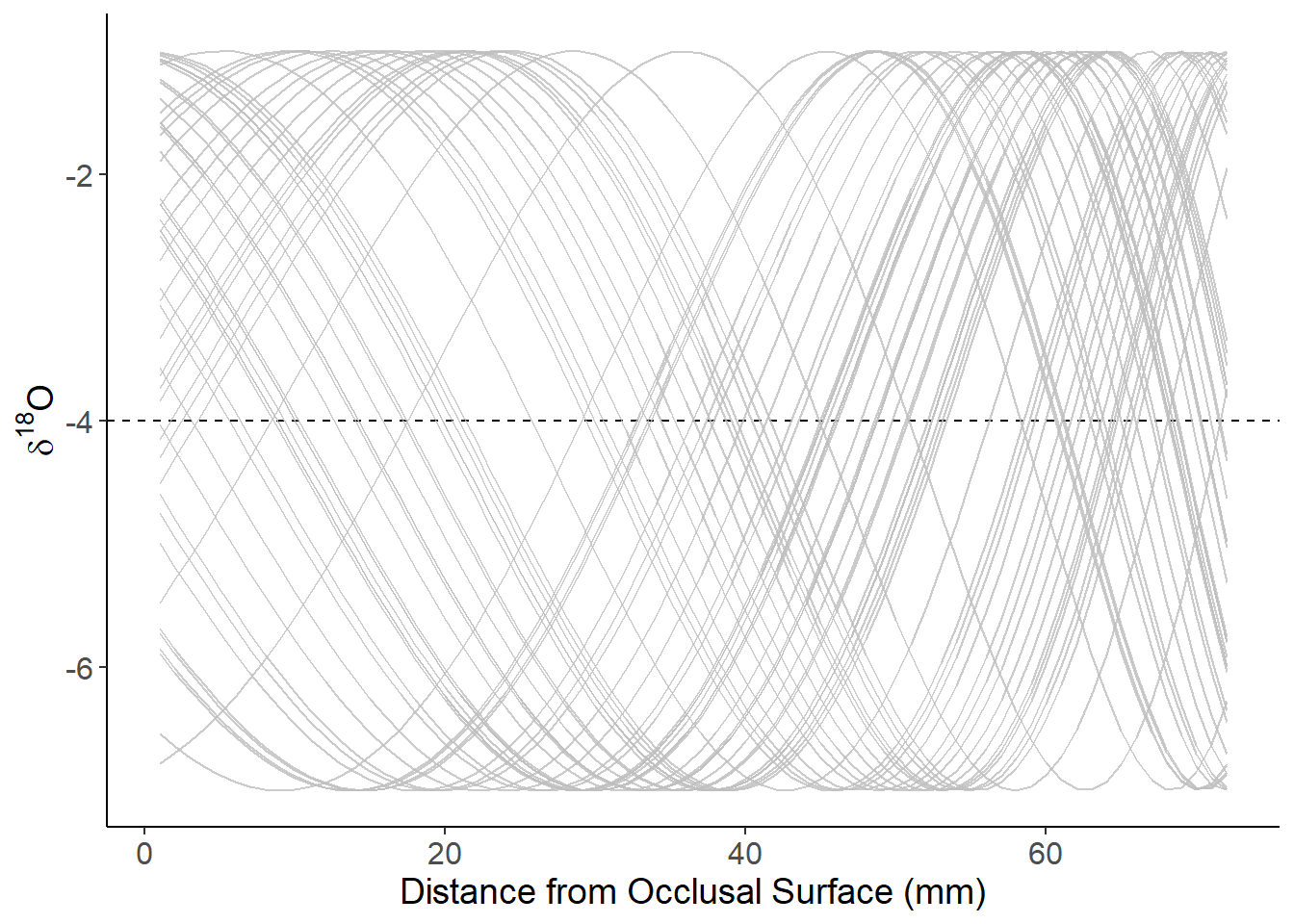
Non-linear growth with variable birth season and death age (Amplitude = 3)
Note: samples chosen random
set.seed(303) #to keep the sampling of teeth consistent
Ex4<-read_csv("simZebraDataEX4.csv") Rows: 33671 Columns: 17
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (1): model
dbl (16): specimen, age_at_death, birth_year, birth_month, birth_day, crown_...
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.Ex4_pivot<-Ex4 %>% pivot_longer(cols=d18O_1:d18O_4,names_to="Amplitude",values_to="d18O",names_prefix="d18O_")
uv<-unique(Ex4$specimen)
samp<-sample(uv,5,replace=FALSE)
Ex4_Amp3_samp<-filter(Ex4_pivot,Amplitude==3,model=="High", specimen %in% samp)
ggplot(Ex4_Amp3_samp,aes(x=mm_band,y=d18O,group=as_factor(specimen),color=as_factor(specimen))) +
geom_hline(yintercept=-4,linetype=2)+
geom_line(lwd=1.1) +
theme_classic() +
scale_color_manual(values=palette.colors(5,palette="R4")) +
labs(x="Distance from Occlusal Surface (mm)",y=expression(paste(delta^"18","O")),color="Specimen #")+
theme(axis.text=element_text(size=12),axis.title=element_text(size=14))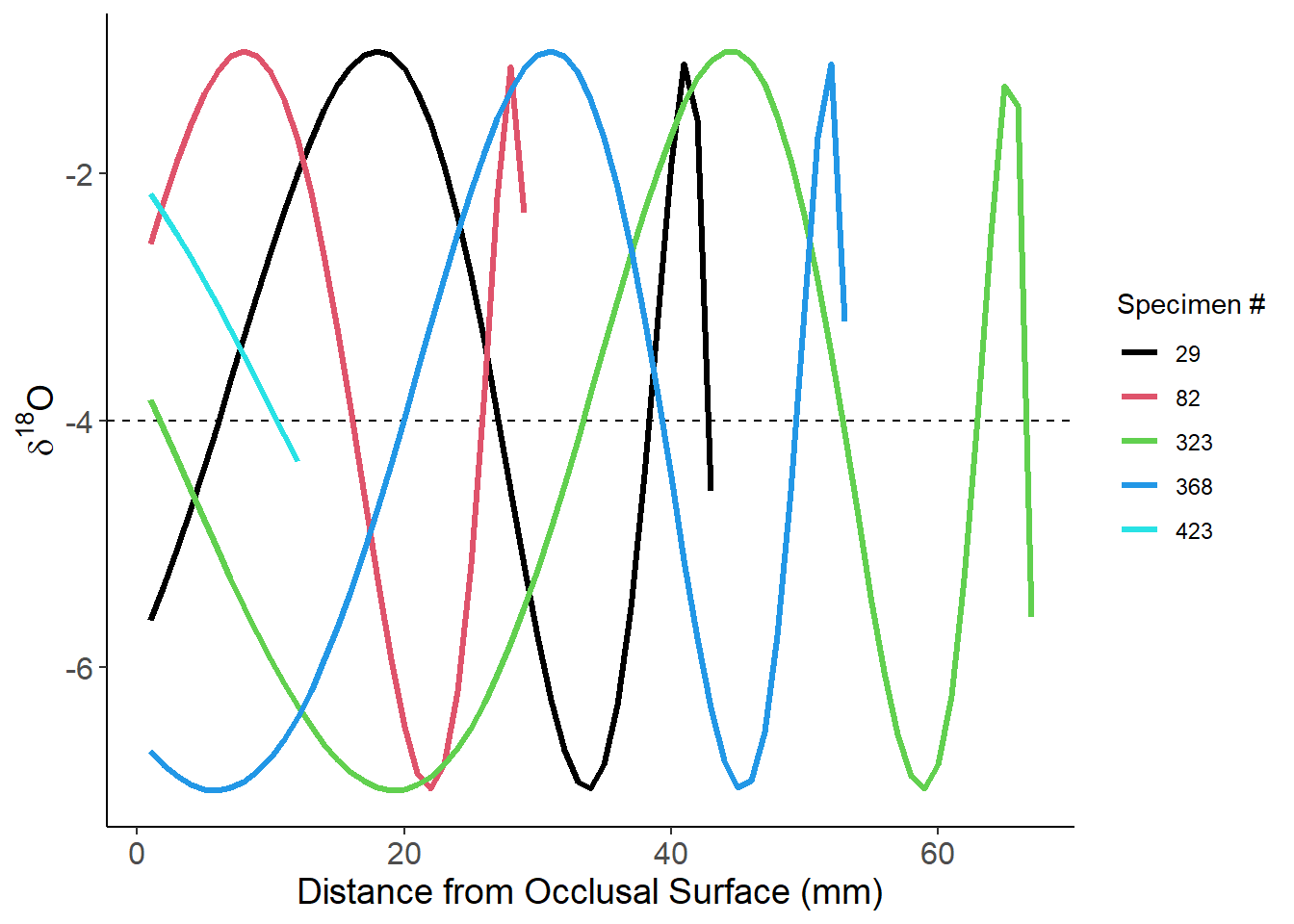
Boxplots of intratooth variation
ggplot(Ex4_Amp3_samp,aes(x=as_factor(specimen),y=d18O,group=as_factor(specimen),color=as_factor(specimen))) +
geom_hline(yintercept=-4,linetype=2)+
geom_boxplot(show.legend = FALSE) +
geom_hline(yintercept=-4,linetype=2)+
theme_classic() +
scale_color_manual(values=palette.colors(5,palette="R4")) +
labs(x="Specimen #",y=expression(paste(delta^"18","O")),color="Specimen #")+
theme(axis.text=element_text(size=12),axis.title=element_text(size=14))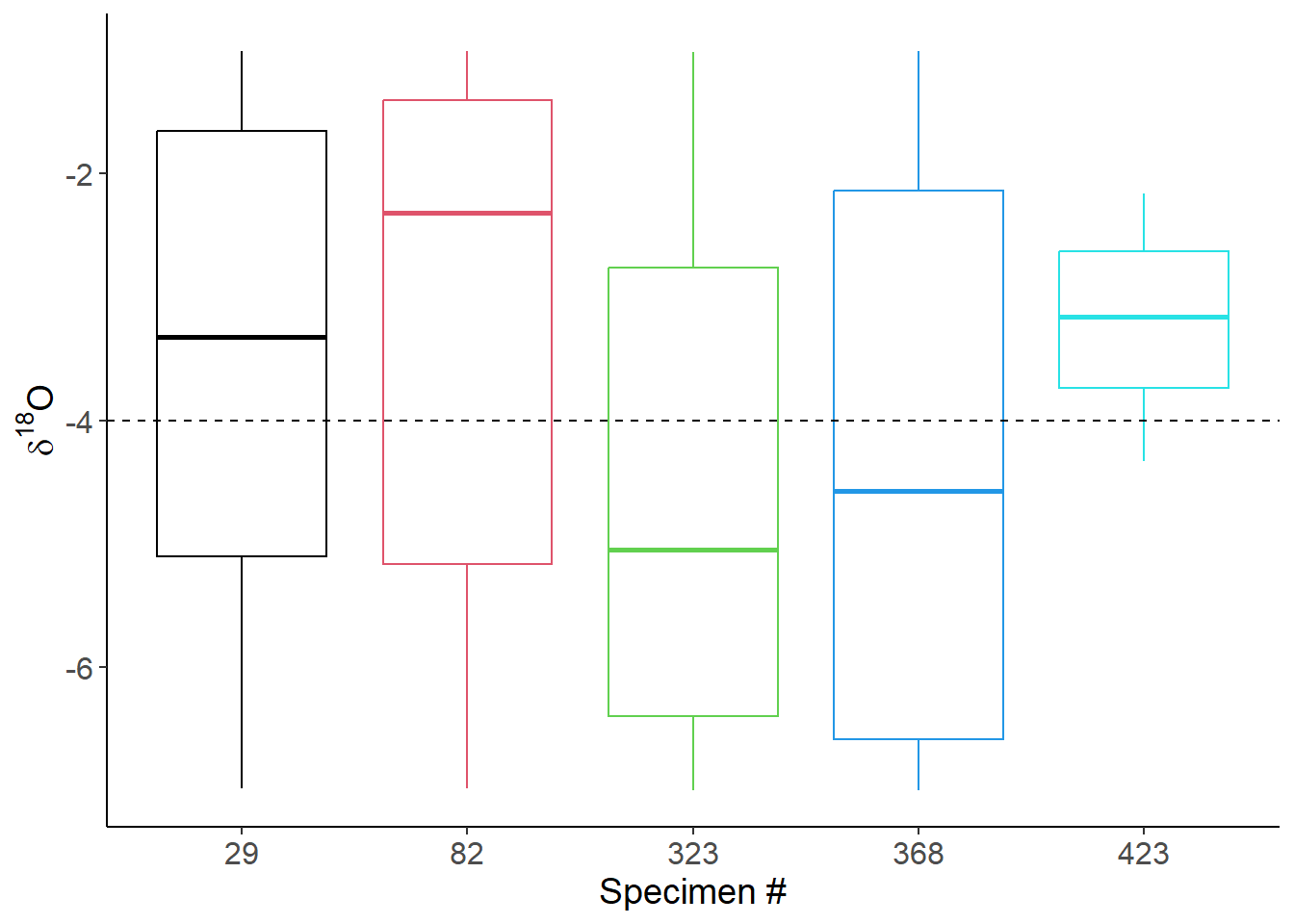
BULK SAMPLING METHOD A: Estimating amplitude from random 1mm bands (Non-linear growth, Amplitude = 3)
Ex4_high<-Ex4 %>% filter(model=="High") %>% drop_na()
sampleSizes<-c(10,20,50,100)
size<-c()
bootstrapAmp<-c()
for (i in sampleSizes) {
for (j in c(1:100)) {
samp<-Ex4_high %>% group_by(specimen) %>% slice_sample(n=1) %>% ungroup() %>% slice_sample(n=i)
size<-append(size,i)
bootstrapAmp<-append(bootstrapAmp,abs(diff(range(samp$d18O_3)))/2)
}
}
data<-data.frame(size,bootstrapAmp)
ggplot(data,aes(x=as_factor(size),y=bootstrapAmp)) +
geom_hline(yintercept=3,linetype=2)+
geom_boxplot() +
theme_classic() +
ylim(c(1,4)) +
labs(x="Sample Size (# of teeth)",y="Estimated Amplitude (\u{2030})") +
theme(axis.text=element_text(size=12),axis.title=element_text(size=14))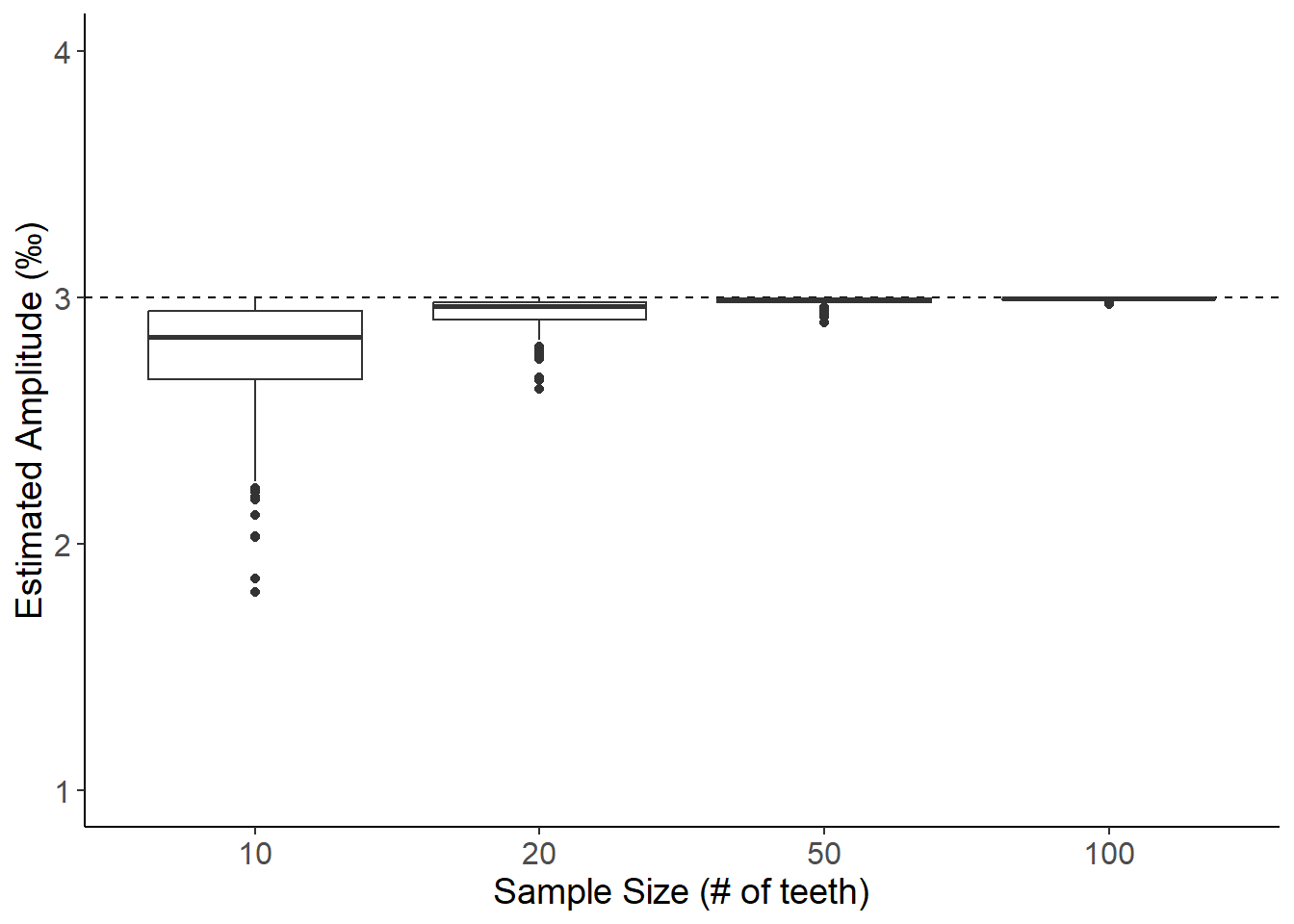
BULK SAMPLING METHOD A: Estimating amplitude from sampling a random 1mm band with 20mm size cutoff (Non-linear growth, Amplitude = 3)
cutoff<-20
Ex4_highCut<-Ex4 %>% filter(model=="High") %>% drop_na() %>% group_by(specimen) %>% filter( n() >=cutoff) %>% ungroup()
sampleSizes<-c(10,20,50,100)
size<-c()
bootstrapAmp<-c()
for (i in sampleSizes) {
for (j in c(1:100)) {
samp<-Ex4_highCut %>% group_by(specimen) %>% slice_sample(n=1) %>% ungroup() %>% slice_sample(n=i)
size<-append(size,i)
bootstrapAmp<-append(bootstrapAmp,abs(diff(range(samp$d18O_3)))/2)
}
}
data<-data.frame(size,bootstrapAmp)
ggplot(data,aes(x=as_factor(size),y=bootstrapAmp)) +
geom_hline(yintercept=3,linetype=2)+
geom_boxplot() +
theme_classic() +
ylim(c(1,4)) +
labs(x="Sample Size (# of teeth)",y="Estimated Amplitude (\u{2030})") +
theme(axis.text=element_text(size=12),axis.title=element_text(size=14))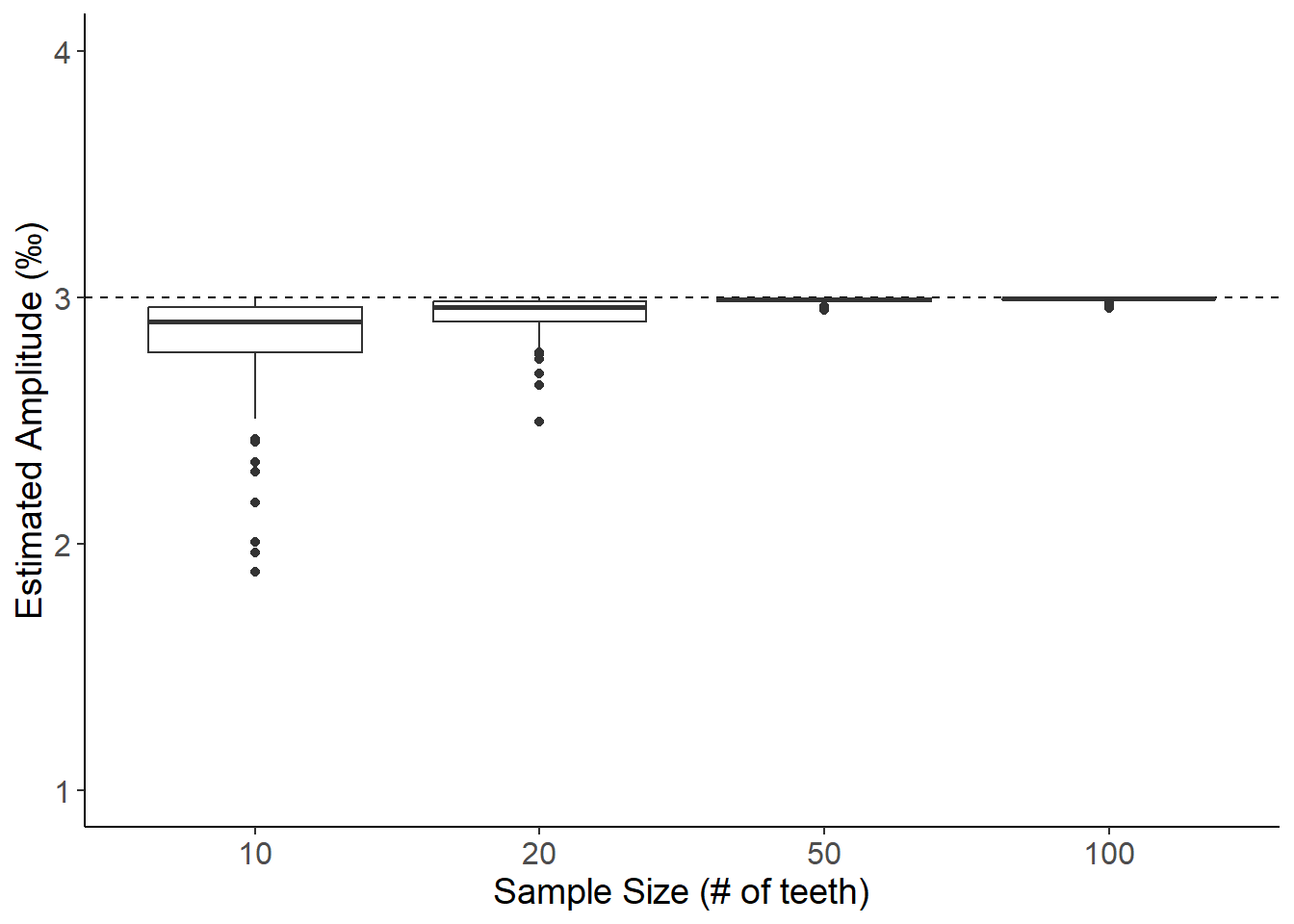
BULK SAMPLING METHOD B: Estimating amplitude from averaged sample taken along growth axis (Non-linear growth, Amplitude = 3)
Ex4_high<-Ex4 %>% filter(model=="High") %>% drop_na()
sampleSizes<-c(10,20,50,100)
size<-c()
bootstrapAmp<-c()
for (i in sampleSizes) {
for (j in c(1:100)) {
samp<-Ex4_high %>% group_by(specimen) %>% slice_sample(n=i) %>% summarize(mean(d18O_3))
size<-append(size,i)
bootstrapAmp<-append(bootstrapAmp,abs(diff(range(samp$`mean(d18O_3)`)))/2)
}
}
data<-data.frame(size,bootstrapAmp)
ggplot(data,aes(x=as_factor(size),y=bootstrapAmp)) +
geom_hline(yintercept=3,linetype=2)+
geom_boxplot() +
theme_classic() +
ylim(c(1,4)) +
labs(x="Sample Size (# of teeth)",y="Estimated Amplitude (\u{2030})") +
theme(axis.text=element_text(size=12),axis.title=element_text(size=14))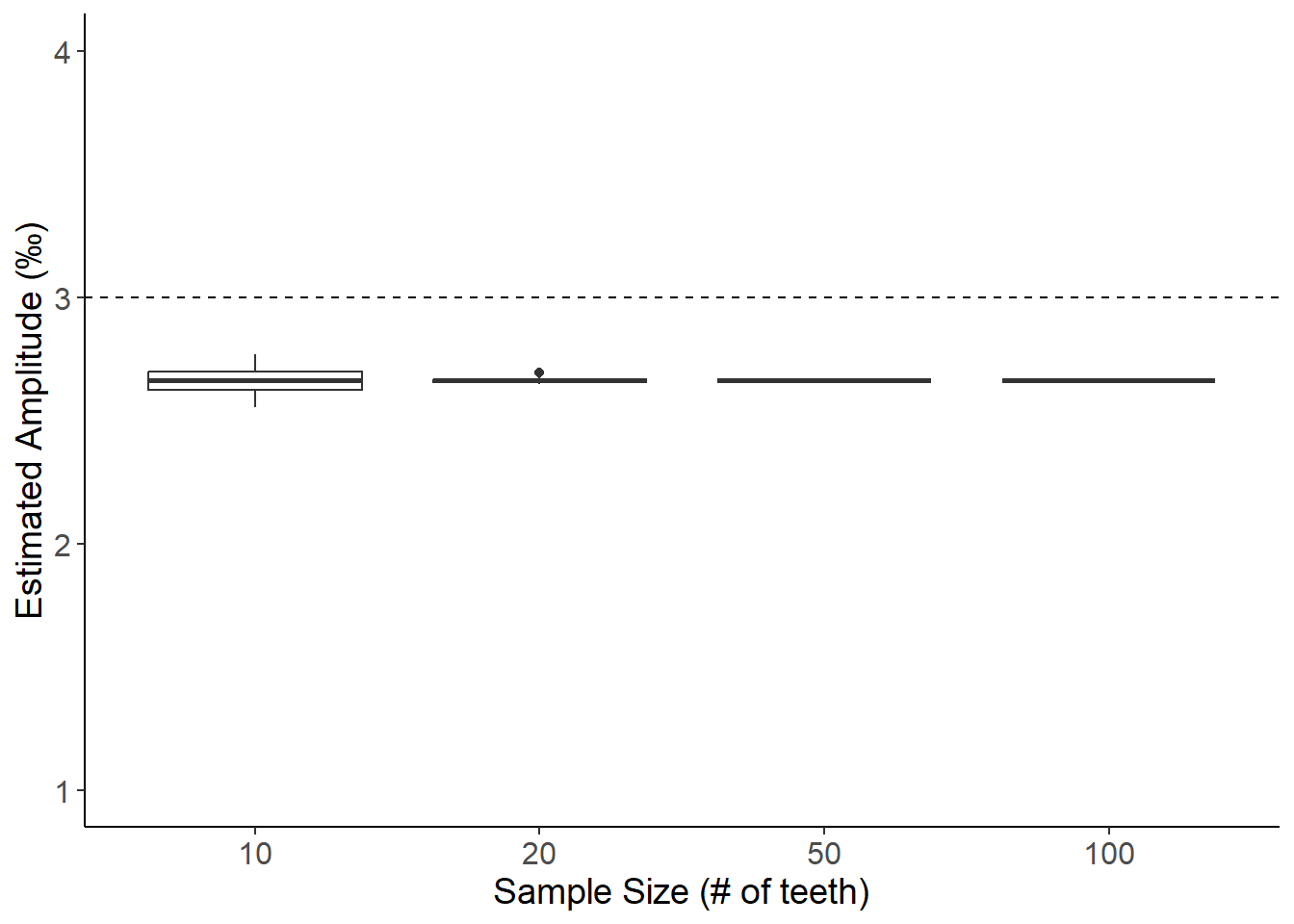
BULK SAMPLING METHOD B: Estimating amplitude from averaged sample taken along growth axis with 20 mm cutoff (Non-linear growth, Amplitude = 3)
cutoff<-20
Ex4_highCut<-Ex4 %>% filter(model=="High") %>% drop_na() %>% group_by(specimen) %>% filter( n() >=cutoff) %>% ungroup()
sampleSizes<-c(10,20,50,100)
size<-c()
bootstrapAmp<-c()
for (i in sampleSizes) {
for (j in c(1:100)) {
samp<-Ex4_highCut %>% group_by(specimen) %>% slice_sample(n=i) %>% summarize(mean(d18O_3))
size<-append(size,i)
bootstrapAmp<-append(bootstrapAmp,abs(diff(range(samp$`mean(d18O_3)`)))/2)
}
}
data<-data.frame(size,bootstrapAmp)
ggplot(data,aes(x=as_factor(size),y=bootstrapAmp)) +
geom_hline(yintercept=3,linetype=2)+
geom_boxplot() +
theme_classic() +
ylim(c(1,4)) +
labs(x="Sample Size (# of teeth)",y="Estimated Amplitude (\u{2030})") +
theme(axis.text=element_text(size=12),axis.title=element_text(size=14))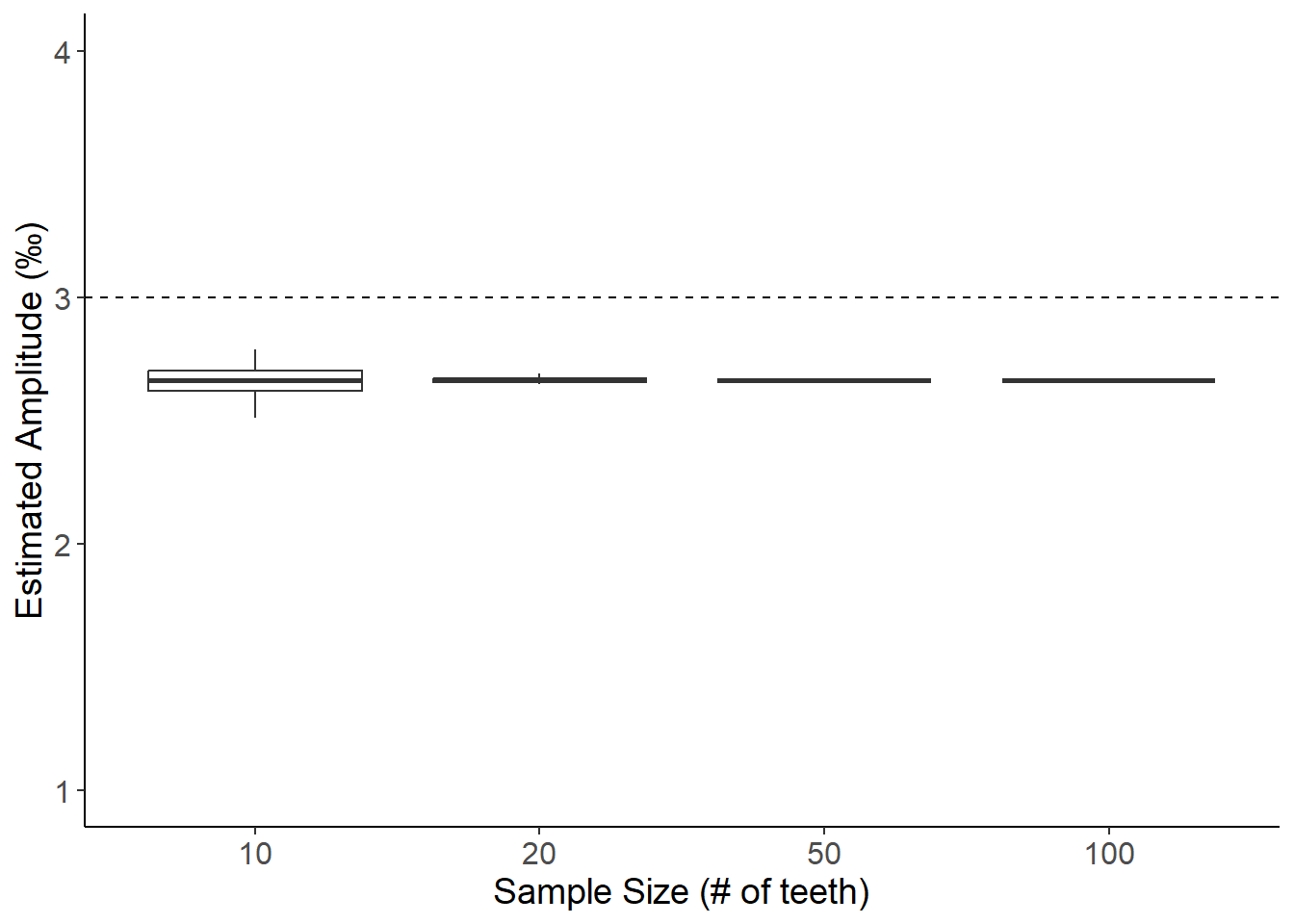
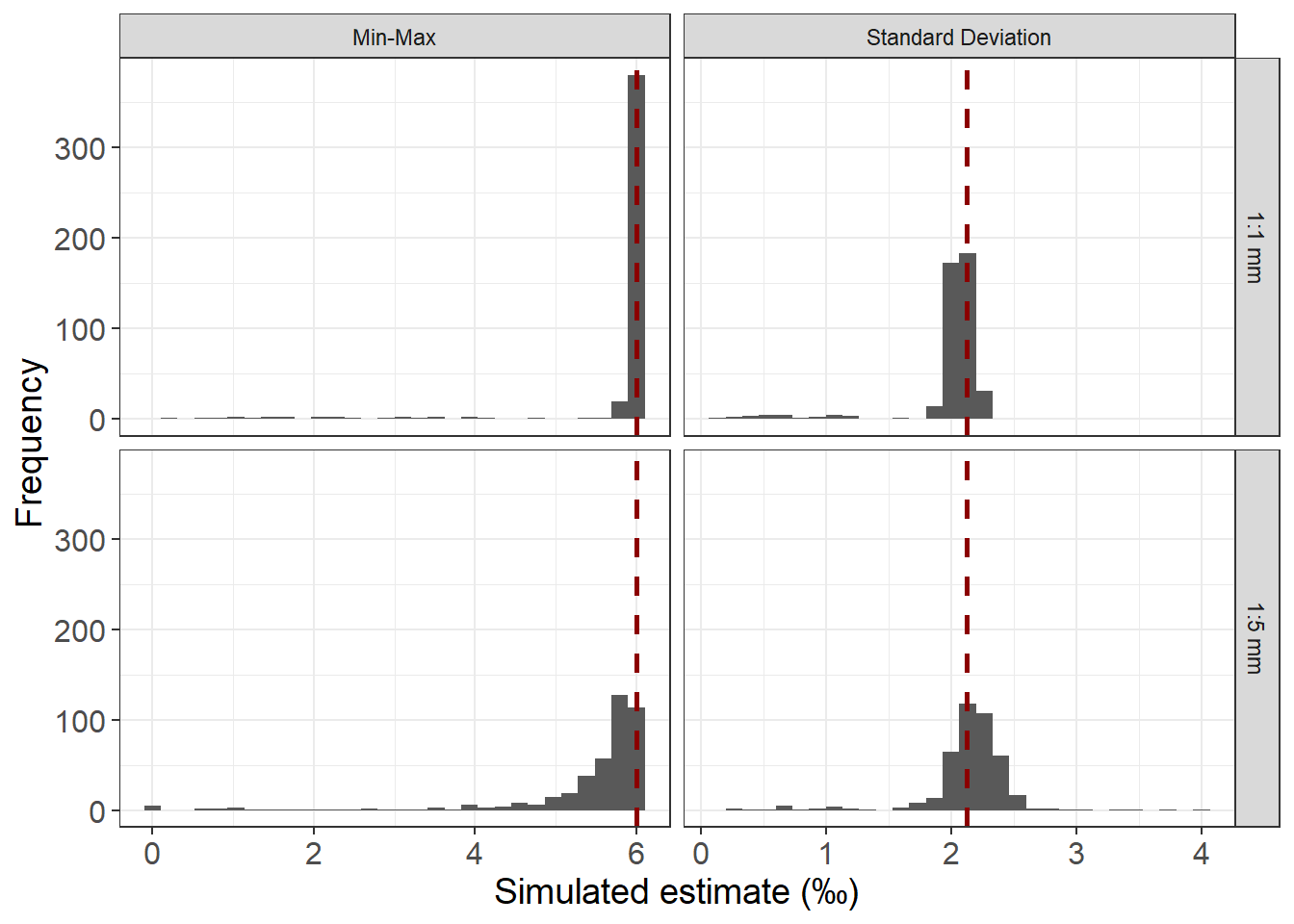
Assessing amplitude
set.seed(303)
ampMethodCheck<-Ex4 %>%
filter(model=="High") %>%
drop_na(d18O_3) %>%
group_by(as_factor(specimen)) %>%
slice_sample(n=200) %>%
arrange(specimen,mm_band) %>%
summarize(stdev=sd(d18O_3),min=min(d18O_3),max=max(d18O_3)) %>%
mutate(minmax=max-min) %>%
rename(specimen=`as_factor(specimen)`)%>%
select(specimen,stdev,minmax) %>%
pivot_longer(
cols=stdev:minmax,
names_to="method",
values_to="value"
) %>%
mutate(samp_freq="1:1 mm")
mids<-rep(c(2.124,6),nrow(ampMethodCheck)/2)
ampMethodCheck<-bind_cols(ampMethodCheck,mids=mids)
ampMethodCheck2<-Ex4 %>%
filter(model=="High") %>%
drop_na(d18O_3) %>%
group_by(as_factor(specimen)) %>%
slice_sample(n=200) %>%
filter(mm_band %% 5 == 0) %>%
arrange(specimen,mm_band) %>%
summarize(stdev=sd(d18O_3),min=min(d18O_3),max=max(d18O_3)) %>%
mutate(minmax=max-min) %>%
rename(specimen=`as_factor(specimen)`)%>%
select(specimen,stdev,minmax) %>%
pivot_longer(
cols=stdev:minmax,
names_to="method",
values_to="value"
)%>%
mutate(samp_freq="1:5 mm")
mids<-rep(c(2.124,6),nrow(ampMethodCheck2)/2)
ampMethodCheck2<-bind_cols(ampMethodCheck2,mids=mids)
ampMethodCheckAll<-bind_rows(ampMethodCheck,ampMethodCheck2) %>%
mutate(method=recode(method,"minmax"="Min-Max","stdev"="Standard Deviation"))
ggplot(ampMethodCheckAll,aes(x=value)) +
geom_histogram() +
geom_vline(aes(xintercept = mids),data=ampMethodCheckAll,lty=2,lwd=1,col="darkred") +
facet_grid(vars(samp_freq),vars(method),scales="free_x") +
theme_bw() +
labs(x="Simulated estimate (\u{2030})",y="Frequency") +
theme(axis.text=element_text(size=12),axis.title=element_text(size=14))`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.Warning: Removed 5 rows containing non-finite outside the scale range
(`stat_bin()`).