stemData<-pivot_longer(
data=treeData,
cols=dbh1:dbh8,
names_to="stemNum",
values_to="dbhMeasure"
)3 Tidying data
In class, we discussed the principles of tidy data:
each variable is stored in its own column
each observation is stored in its own row
each cell contains a unique value
These seem pretty straightforward, but in fact these simple rules are broken quite regularly, with data stored in column names, or multiple data types stored in a single column, or variables stored in both rows and columns. The tidyr package provides some key functions for dealing with these kinds of issues.
3.0.1 Pivot_longer
Oftentimes, categorical information you want to use is actually stored in the names of columns. To deal with this, we can gather the data in these columns into two columns: one with the column names, and another with their associated values.
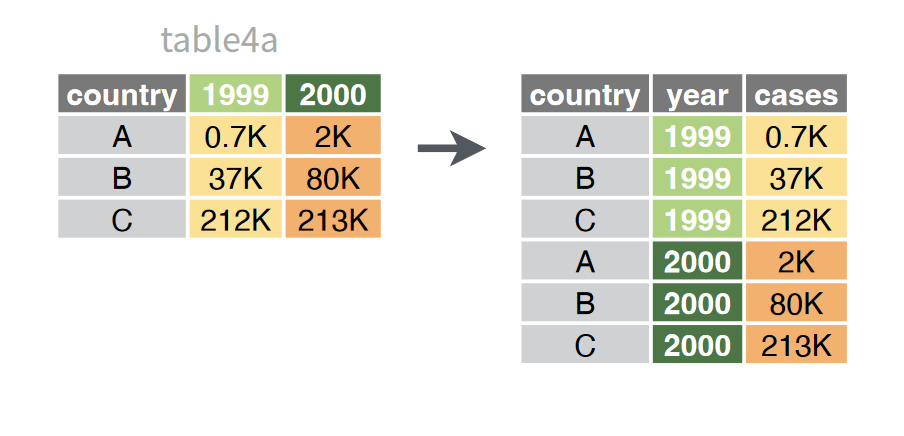
In the tree dataset, for example, the last 8 columns are values for DBH recorded across multiple stems (up to 8). Not every tree will have these, so the table uses -1 to indicate an absence of stems. We can use pivot_longer to turn each of these columns into a value in a single column:
Here’s a breakdown of the arguments being used here:
datais the table being pivoted (in this case, treeData)colsis the columns where the names are going to be converted to data (in this case, columns dbh1 to dbh8 (you could substitute a vector of columns names if they aren’t in sequential order)names_toprovides a name for a new column where the names are going to be stored as a datavalues_toprovides a name for a new column where the values from the columns we are converting will go.
Running this code, you can see the effect. While we’re at it, we might want to eliminate the letters “dbh” from the stem number values. We can do this by adding an additional argument: names_prefix.
stemData<-pivot_longer(
data=treeData,
cols=dbh1:dbh8,
names_to="stemNum",
values_to="dbhMeasure",
names_prefix = "dbh"
)Now when we look at our data, we have a few less variables to deal with but a considerably larger number of observations:
stemData# A tibble: 115,896 × 35
DbaseID Region City Source TreeID Zone `Park/Street` SpCode ScientificName
<dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr> <chr>
1 1 InlVal Modes… Motow… 1 Nurs… Nursery ACSA1 Acer sacchari…
2 1 InlVal Modes… Motow… 1 Nurs… Nursery ACSA1 Acer sacchari…
3 1 InlVal Modes… Motow… 1 Nurs… Nursery ACSA1 Acer sacchari…
4 1 InlVal Modes… Motow… 1 Nurs… Nursery ACSA1 Acer sacchari…
5 1 InlVal Modes… Motow… 1 Nurs… Nursery ACSA1 Acer sacchari…
6 1 InlVal Modes… Motow… 1 Nurs… Nursery ACSA1 Acer sacchari…
7 1 InlVal Modes… Motow… 1 Nurs… Nursery ACSA1 Acer sacchari…
8 1 InlVal Modes… Motow… 1 Nurs… Nursery ACSA1 Acer sacchari…
9 2 InlVal Modes… Motow… 2 Nurs… Nursery BEPE Betula pendula
10 2 InlVal Modes… Motow… 2 Nurs… Nursery BEPE Betula pendula
# ℹ 115,886 more rows
# ℹ 26 more variables: CommonName <chr>, TreeType <chr>, address <chr>,
# street <chr>, side <chr>, cell <dbl>, OnStreet <chr>, FromStreet <chr>,
# ToStreet <chr>, Age <dbl>, `DBH (cm)` <dbl>, `TreeHt (m)` <dbl>,
# CrnBase <dbl>, `CrnHt (m)` <dbl>, `CdiaPar (m)` <dbl>,
# `CDiaPerp (m)` <dbl>, `AvgCdia (m)` <dbl>, `Leaf (m2)` <dbl>,
# Setback <dbl>, TreeOr <dbl>, CarShade <dbl>, LandUse <dbl>, Shape <dbl>, …What this data allows us to do more easily is use stem number itself as a variable. For example, we can now plot a boxplot to show how these vary in terms of DBH measurements:
stemData2<-filter(stemData,dbhMeasure>-1)
ggplot(stemData2,aes(x=stemNum,y=dbhMeasure)) +
geom_boxplot()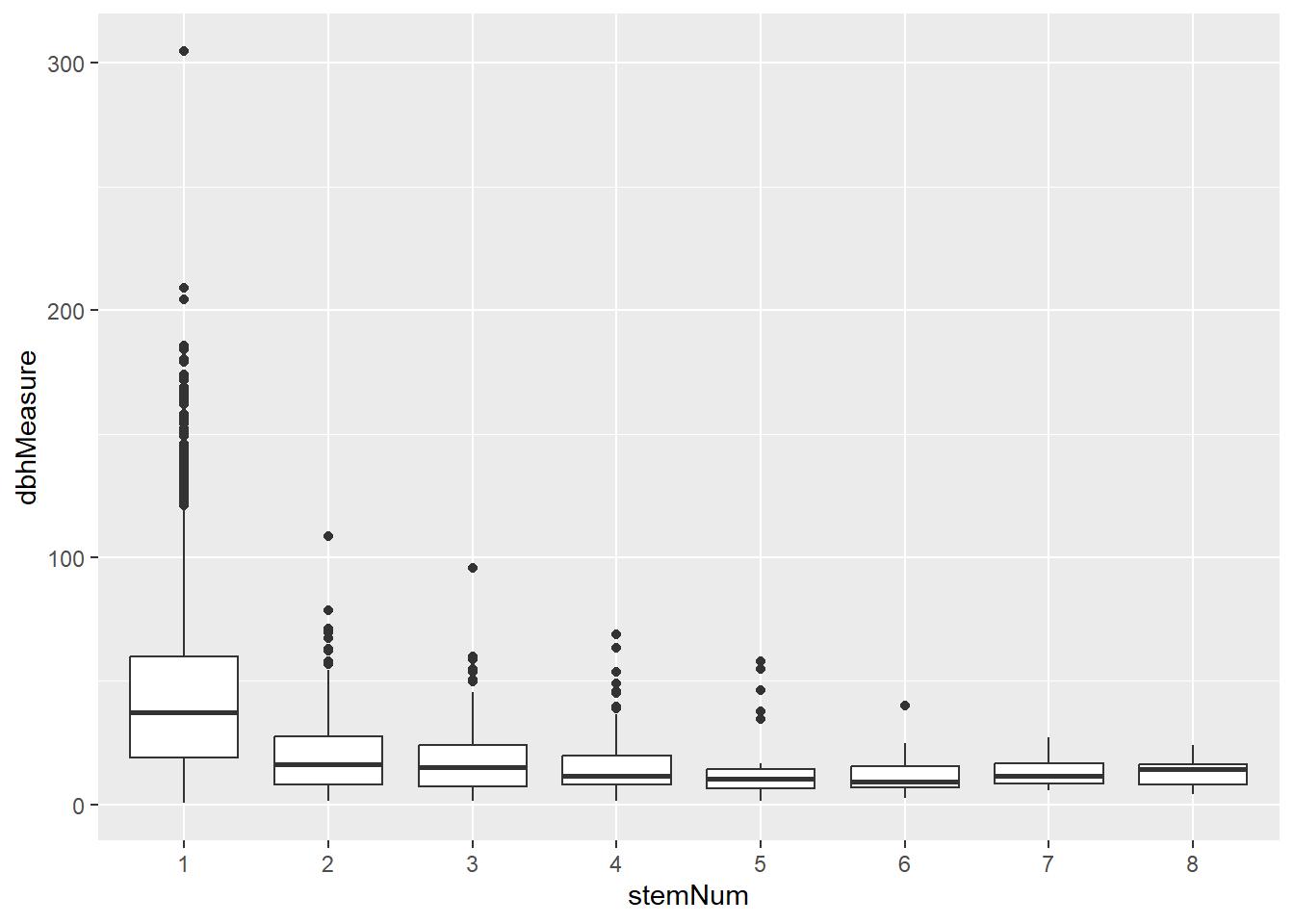
3.0.2 Pivot_wider
Pivoting wider involves taking data that is stored in a single column and spreading it across multiple columns.
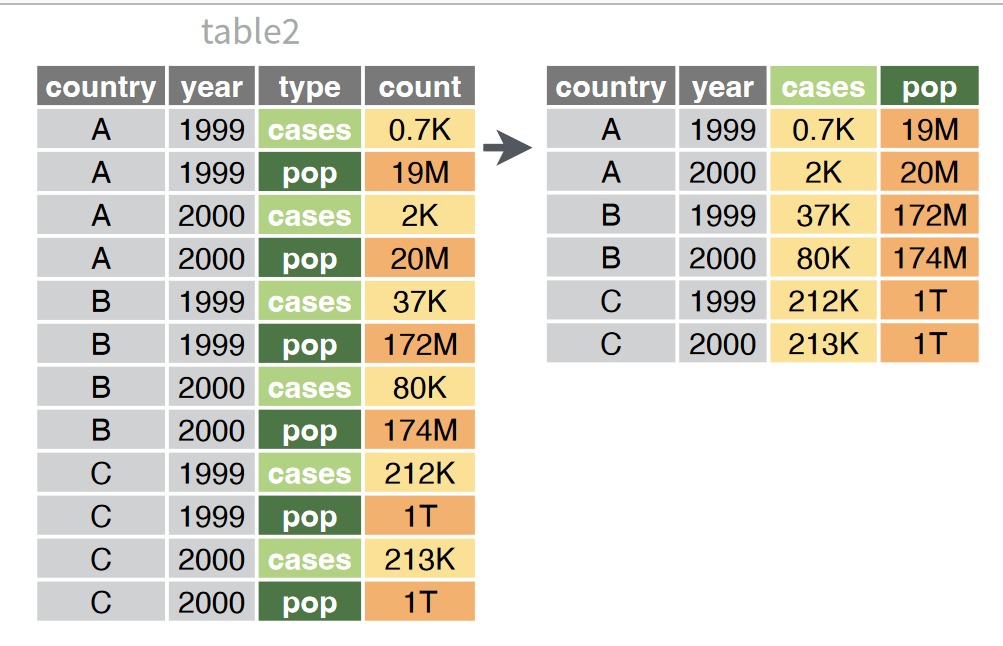
Here, we’ll use it to convert out tree data back into the format we’re used to:
treeWide<-pivot_wider(
data=stemData,
names_from=stemNum,
values_from=dbhMeasure
)
treeWide# A tibble: 14,487 × 41
DbaseID Region City Source TreeID Zone `Park/Street` SpCode ScientificName
<dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr> <chr>
1 1 InlVal Modes… Motow… 1 Nurs… Nursery ACSA1 Acer sacchari…
2 2 InlVal Modes… Motow… 2 Nurs… Nursery BEPE Betula pendula
3 3 InlVal Modes… Motow… 3 Nurs… Nursery CESI4 Celtis sinens…
4 4 InlVal Modes… Motow… 4 Nurs… Nursery CICA Cinnamomum ca…
5 5 InlVal Modes… Motow… 5 Nurs… Nursery FRAN_R Fraxinus angu…
6 6 InlVal Modes… Motow… 6 Nurs… Nursery FREX_H Fraxinus exce…
7 7 InlVal Modes… Motow… 7 Nurs… Nursery FRHO Fraxinus holo…
8 8 InlVal Modes… Motow… 8 Nurs… Nursery FRPE_M Fraxinus penn…
9 9 InlVal Modes… Motow… 9 Nurs… Nursery FRVE_G Fraxinus velu…
10 10 InlVal Modes… Motow… 10 Nurs… Nursery GIBI Ginkgo biloba
# ℹ 14,477 more rows
# ℹ 32 more variables: CommonName <chr>, TreeType <chr>, address <chr>,
# street <chr>, side <chr>, cell <dbl>, OnStreet <chr>, FromStreet <chr>,
# ToStreet <chr>, Age <dbl>, `DBH (cm)` <dbl>, `TreeHt (m)` <dbl>,
# CrnBase <dbl>, `CrnHt (m)` <dbl>, `CdiaPar (m)` <dbl>,
# `CDiaPerp (m)` <dbl>, `AvgCdia (m)` <dbl>, `Leaf (m2)` <dbl>,
# Setback <dbl>, TreeOr <dbl>, CarShade <dbl>, LandUse <dbl>, Shape <dbl>, …3.0.3 Separate
The seperate function takes columns where there is more than one piece of information and breaks it up into multiple columns. For example, let’s say we wanted separate columns for the genus and species of the trees:
treeDataSep<-separate(
data=treeData,
col=ScientificName,
into=c("Genus","Species"),
sep=" ",
extra="drop")
treeDataSep# A tibble: 14,487 × 42
DbaseID Region City Source TreeID Zone `Park/Street` SpCode Genus Species
<dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr> <chr> <chr>
1 1 InlVal Modest… Motow… 1 Nurs… Nursery ACSA1 Acer saccha…
2 2 InlVal Modest… Motow… 2 Nurs… Nursery BEPE Betu… pendula
3 3 InlVal Modest… Motow… 3 Nurs… Nursery CESI4 Celt… sinens…
4 4 InlVal Modest… Motow… 4 Nurs… Nursery CICA Cinn… campho…
5 5 InlVal Modest… Motow… 5 Nurs… Nursery FRAN_R Frax… angust…
6 6 InlVal Modest… Motow… 6 Nurs… Nursery FREX_H Frax… excels…
7 7 InlVal Modest… Motow… 7 Nurs… Nursery FRHO Frax… holotr…
8 8 InlVal Modest… Motow… 8 Nurs… Nursery FRPE_M Frax… pennsy…
9 9 InlVal Modest… Motow… 9 Nurs… Nursery FRVE_G Frax… veluti…
10 10 InlVal Modest… Motow… 10 Nurs… Nursery GIBI Gink… biloba
# ℹ 14,477 more rows
# ℹ 32 more variables: CommonName <chr>, TreeType <chr>, address <chr>,
# street <chr>, side <chr>, cell <dbl>, OnStreet <chr>, FromStreet <chr>,
# ToStreet <chr>, Age <dbl>, `DBH (cm)` <dbl>, `TreeHt (m)` <dbl>,
# CrnBase <dbl>, `CrnHt (m)` <dbl>, `CdiaPar (m)` <dbl>,
# `CDiaPerp (m)` <dbl>, `AvgCdia (m)` <dbl>, `Leaf (m2)` <dbl>,
# Setback <dbl>, TreeOr <dbl>, CarShade <dbl>, LandUse <dbl>, Shape <dbl>, …The arguments here should look familiar, but a couple may be worth discussing. The sep argument gives the symbol that separates the two sections. Here, the genus and species are separated by a single space, so we include that in quotes. The extra argument gives instructions on what to do with any extra data. If you look at the original tree data, you can see some scientific names have more than two parts. In this case, let’s assume we’re not too fussy about that, so we use the argument “drop” to get rid of it.
3.0.4 Unite
Unite is the opposite of separate. It simply takes two columns and puts them together, like so:
treeDataUnite<-unite(
data=treeDataSep,
col="SciName",
Genus:Species,
sep=" ",
remove=TRUE)
treeDataUnite# A tibble: 14,487 × 41
DbaseID Region City Source TreeID Zone `Park/Street` SpCode SciName
<dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr> <chr>
1 1 InlVal Modesto, CA Motown2… 1 Nurs… Nursery ACSA1 Acer s…
2 2 InlVal Modesto, CA Motown2… 2 Nurs… Nursery BEPE Betula…
3 3 InlVal Modesto, CA Motown2… 3 Nurs… Nursery CESI4 Celtis…
4 4 InlVal Modesto, CA Motown2… 4 Nurs… Nursery CICA Cinnam…
5 5 InlVal Modesto, CA Motown2… 5 Nurs… Nursery FRAN_R Fraxin…
6 6 InlVal Modesto, CA Motown2… 6 Nurs… Nursery FREX_H Fraxin…
7 7 InlVal Modesto, CA Motown2… 7 Nurs… Nursery FRHO Fraxin…
8 8 InlVal Modesto, CA Motown2… 8 Nurs… Nursery FRPE_M Fraxin…
9 9 InlVal Modesto, CA Motown2… 9 Nurs… Nursery FRVE_G Fraxin…
10 10 InlVal Modesto, CA Motown2… 10 Nurs… Nursery GIBI Ginkgo…
# ℹ 14,477 more rows
# ℹ 32 more variables: CommonName <chr>, TreeType <chr>, address <chr>,
# street <chr>, side <chr>, cell <dbl>, OnStreet <chr>, FromStreet <chr>,
# ToStreet <chr>, Age <dbl>, `DBH (cm)` <dbl>, `TreeHt (m)` <dbl>,
# CrnBase <dbl>, `CrnHt (m)` <dbl>, `CdiaPar (m)` <dbl>,
# `CDiaPerp (m)` <dbl>, `AvgCdia (m)` <dbl>, `Leaf (m2)` <dbl>,
# Setback <dbl>, TreeOr <dbl>, CarShade <dbl>, LandUse <dbl>, Shape <dbl>, …The last argument in this case, remove, takes away the now redundant columns of Genus and Species.
Use the separate and unite functions to do the following:
In the tree dataset, create separate columns for city and state
In the Sacramento dataset, combine the latitude and longitude columns